Screening and identification of purine degrading Lactobacillus fermentum 9-4 from Chinese fermented rice- flour noodles
Lianghua Lu, Tiantian Liu, Xiaoling Liu, Chenghua Wang*
College of Light Industry and Food Engineering, Guangxi University, Nanning 530004, China
Keywords:
Hyperuricemia
Probiotics
Lactobacillus fermentum
Low-purine food
Fermented rice noodle
A B S T R A C T
The aim of this study was to isolate a new purine-degrading potential probiotic strain from Chinese fermented rice- flour noodles and investigate its potential application in purine-degrading food development for promising anti-gout therapy. A new lactic acid bacteria strain designated as Lactobacillus fermentum 9-4 was screened out from 10 Shengzhamifen samples by a comprehensive method integrating agar plate selection, in vitro purine-metabolizing enzymatic activities of xanthine oxidase and urate oxidase, 16S rRNA gene sequencing and identification. The resting cells of L. fermentum 9-4 showed the maximum degradation rates of inosine and guanosine by respective 2.13 × 10-3 and 2.78 × 10-3 g/(L·min), and the highest assimilation ratio of guanosine by (55.93 ± 3.12)%, which are improvements over LAB strains characterized previously. Yogurt fermented by L. fermentum 9-4 also efficiently assimilated the inosine and guanosine, with respective degradation rates of 98.10% and 98.56% higher than those of the commercial ones. The L. fermentum 9-4 showed excellent survival (> 80%) under the conditions of pH 2.5 and 0.1% bile salt. The results suggest that L. fermentum 9-4 may be a promising candidate as a probiotic for developing low-purine foods.
1. Introduction
Hyperuricemia describes the abnormally high level of uric acid in the blood caused by excessive production and reduced excretion of uric acid, and eventually develops into gout [1]. Hyperuricemia is also a risk factor for chronic kidney disease [2], diabetes [3],cardiovascular disease [4]and other metabolic syndrome [5].Epidemiological investigations showed that the prevalence rate of hyperuricemia is about 20% in the USA, and the situation is even more serious in the southeast coastal areas of China [6,7]. So more and more attention has been paid to the control of hyperuricemia and gout.
Up to now, two approaches have been developed to treat hyperuricemia and gout: pharmacotherapy and dietotherapy [1,8]. The former employs chemical medicines to improve symptoms, reduce uric acid production, promote uric acid excretion or decomposition with some inevitable side effects, like gastrointestinal bleeding and other symptoms [9], which disables it a long-term solution, especially for asymptomatic hyperuricemia [10]. In contrast, the latter relies on a strict low-purine diet to control purine intake, which reduces uric acid level in food during consumption, resulting in the reduction of uric acid in the blood without any toxic side effects [11]. However,strict dietary restrictions exclude the intake of nutritional purine-rich foods including most meat, seafood, and some vegetables, which is not conducive to nutritional balance. Meanwhile, in our daily lives,the compliance to strict dietary restrictions is usually very low due to two reasons: lack of accurate information of purine content in various foods consumed; poor palatability of the low-purine foods due to the deficiency of flavor compound purines like inosine monophosphate acid (IMP) and guanosine monophosphate acid (GMP) [12]. A very promising and creative way may be developing dietary supplements like probiotics that can assimilate or degrade purines of nutritional purine-rich foods in food ingestion and consumption processes to reduce purine absorption in human intestine, that mainly determines the serum uric acid level [13,14].
Lactic acid bacteria (LAB) are generally recognized as safe (GRAS)and have been used as probiotics for anti-inflammatory effects [15],nutrient metabolism [16,17]and adjustment of the structure of intestinal flora [18]. Recently, a few LAB strains were reported to be involved in purine metabolism and degradation, indicating potential benefits in prevention and treatment of hyperuricemia and gout. The strains include a purine-nucleotide-degradingLactobacillus plantarumDM9218-A [19]isolated from Chinese Sauerkraut, a purine-reducingL. gasseristrains PA-3 [20]issued by Meiji Dairy and a guanosine-degradingL. fermentumisolated by Hangzhou Wahaha Technology Co., Ltd., China [21]. The level of serum uric acid in hyperuricemic rat can be efficiently reduced by the intragastric administration ofL. plantarumDM9218-A with purine degradation abilityin vitro, and the absorption of purines compounds was reduced by oral treatment ofL. gasseriPA-3. However, all these strains mainly degrade the purine nucleosides and nucleotides to purine bases, which can finally be absorbed by the small intestine and further metabolized into uric acid contributing to the hyperuricemia and gout. Moreover, hardly any simultaneousin vivoandin vitrostudies of purine metabolizing enzymes of probiotics and their purine assimilation ability have been reported. These necessitate developing novel purine probiotics with higher purine nucleoside assimilation and purine metabolizing and degrading abilities.
Shengzhamifen, an extruding rice noodles, is a traditional Chinese fermented rice-flour noodles with the characteristics of Guangxi Zhuang Autonomous Region. It is believed that the nutritional value, functionality and specialty can be partially attributed to the prerequisite fermentation of the rice flour by the probiotics including LAB adaptable to local weather conditions. However, few attentions have been paid on LAB from these Chinese fermented rice-flour noodles. The previously reported screening of purine nucleoside degrading LAB strains from plant-based fermented foods like Chinese Sauerkraut inspires us to try to do the same thing with fermented rice-flour noodle, although no reasonable relationship between the functionality of purine-degrading and Shengzhamifen has ever been reported before. This study reported the unexpected finding of new purine-degradingL. fermentum9-4 firstly screened from Shengzhamifen and its evaluation on potential application in purine-degrading food development for promising anti-gout therapy.
2. Materials and methods
2.1 Materials
Ten samples of Shengzhamifen fermented flours were collected from ten old-established shops with characteristics of Zhuang nationality in Nanning City of Guangxi Zhuang Autonomous Region of China. The sample information is listed in Table 1. MRS medium was used to isolate and culture LAB strains. PrimeSTARTMDNA polymerase was purchased from China Dalian Takara Bio Co., Ltd.DNA Cycle Pure Kit was acquired from America Omega Co., Ltd.Super-Bradford Protein Assay Kit was purchased from Beijing Comwin Biotech Co., Ltd. (China). Xanthine, inosine and guanosine were purchased from Sigma Company (Germany) and bovine bile salt was from Beijing Solarbio Science & Technology Co., Ltd. (China).All the other chemicals used in this study were analytical grade and commercially available.

Table 1Source, pH and viable bacterial count of 10 Shengzhamifen samples used in this study.
2.2 Screening and identification procedures of purine-degrading LAB
2.2.1 Agar plate selection
2.5 g of Shengzhamifen fermented flour were dissolved in 22.5 mL sterilized normal saline as 10-1dilution, 10-1dilution was diluted 10 times with normal saline as 10-2dilution, repeat the process until 10-7dilution. 0.1 mL aliquots of each dilution (10-1, 10-3, 10-5and 10-7)were inoculated in triplicate on MRS agar containing 2% CaCO3and 0.01 mg/mL nystatin (preventing mold pollution) by spread-plate method. After incubation at 30 °C for 48 h, colonies with LAB morphology according to the “Bergey’s Manual of Systematic Bacteriology” and calcium dissolving circle were chosen. A total of 35 candidate strains were obtained from about 6 000 colonies,and designated sequentially as 1-1-1-4, 2-1-2-4, 4-1-4-3, 5-1-5-4,6-1-6-3, 7-1-7-3, 8-1-8-3, 9-1-9-4, 10-1-10-3.
2.2.2 Screening by in vitro purine-metabolizing enzymatic activities of xanthine oxidase (XOD) and urate oxidase (UOX)
The candidate strains were cultured in MRS at 30 °C for 24 h,respectively. Three milliliters of cultures were centrifuged at 4 °C,10 000 r/min for 1 min, and the precipitated cells were suspended in 100 μL 50 mmol/L phosphate buffer solution (PBS, pH 7.5). The suspension was ground at -30 °C for 240 s in a high-throughput tissue grinder SCIENTZ-48L (Ningbo Scientz Biotechnology Co.,Ltd., China) and then centrifuged at 4 °C, 13 000 r/min for 2 min.The cell-free supernatants were detected for enzymatic activities of XOD and UOX by spectrometry methods according to our previous works [22,23], protein concentration was determined by the method of Bradford. All samples were analyzed in triplicate. One XOD unit was defined as the amount of enzyme that converts one micro mole of xanthine into uric acid per minute at room temperature and pH 8.5,and that of UOX was the amount of enzyme to convert one micro mole of uric acid into allantoin under the same assay conditions.Strains showing obvious XOD and UOX activities were chosen for the next round of screening.
2.2.3 Identification by 16S rDNA sequencing
After cultured in MRS medium at 30 °C for 24 h, 4.5 mL culture of each chosen strains was used to extract genome DNA with a bacterial genome DNA extraction kit. By using the extracted genomic DNA as template, a pair of universal primers: 27F, 5’-AGAGTTTGATCCTGGCTCAG-3’ and 1492R,5’-GGTTACCTTGTTACG ACTT-3’, and PrimeSTARTMDNA polymerase, 16S rDNA of each strain was PCR amplified. PCR products were purified by using DNA product purification kit and were sequenced at Liuhe Huada Gene Technology Co.,Ltd. (Guangzhou, China). The 16S rDNA sequences of selected strains were analyzed by BLAST in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the RDP database (http://RDP.cme.msu.edu/). The phylogenetic relationships between strains were analyzed by MEGA-X [24]. The neighbor-joining tree [25]was drawn with bootstrap probabilities determined using 1 000 replicates and presented as the percentage [26]. The LAB strains were chosen as candidates for further screening.
2.2.4 Screening of purine-nucleoside-degrading LAB by in vivo degrading abilities on inosine and guanosine
The candidate LAB strains were further screened based on theirin vivopurine degrading abilities on inosine and guanosine by using resting cells. After cultivated in MRS medium at 30 °C for 24 h,2 mL of each culture was centrifuged at 4 °C, 12 000 r/min for 3 min and the precipitated cells was washed twice with normal saline (0.9% NaCl), and then resuspended in 1 mL of 50 mmol/L PBS (pH 7.5) to make the resting cells. The resting cells were transformed into 2 mL of inosine-guanosine solution (each 1 mmol/L in 50 mmol/L pH 7.5 PBS) to start the purine nucleotide degradation reaction and incubated at 160 r/min, 37 °C for 60 min. After incubation, the reaction solution was centrifuged at 4 °C, 12 000 r/min for 3 min, and 2 mL of supernatant was taken out to inactivate enzyme by heating at 90 °C for 20 min. After filtration (0.22 μm), 20 μL permeate was used for determining the inosine and guanosine contents by HPLC analysis according to previous study with minor revisions stated below [27].All experiments were performed in triplicate. The degrading speed and ratio values of inosine and guanosine by each LAB strain were calculated according to the following formula:

V: degradation speed (mg/(L·min)),Mr: Relative molecular mass of inosine or guanosine,n: content of inosine or guanosine before degradation (mmol),X: content of inosine or guanosine (mmol) after degradation,α: degradation ratio.
HPLC analysis for nucleoside contents was carried out at 35 °C with an Eclipse Plus C18column (4.6 mm × 250 mm, 5 μm,Agilent, USA) in a HPLC device equipped with a 2998 photodiode array detector (Waters e2695, USA), and UV-absorbance spectra at 243 nm were recorded. The mobile phase was CH3OH-NaH2PO4solution(methanol:sodium phosphate (10 mmol/L, pH 4.7) = 1:9) at a flow rate of 0.5 mL/min. The reference standards of inosine and guanosine were purchased from Sigma Company (Germany). The standard curves between content and peak area for inosine and guanosine were respective:

A: the peak area,n: amount of substance (mmol).
2.3 Acid and bile salt tolerance test
For the acid resistance test, 1 mL overnight MRS broth cultures ofL. fermentum9-4 were centrifuged at 4 °C, 8 000 r/min for 5 min,the precipitate cells were washed twice with 1 mL of 0.9% saline salt and then inoculated into 1 mL MRS broth acidified to pH 1.0, 2.0 or 3.0 with 1 mol/L hydrochloric acid. The broths were cultured at 37 °C and the survival at 1, 2, and 3 h was ascertained in MRS agar after incubation at 37 °C for 24 h.
Bile salt tolerance ofL. fermentum9-4 was ascertained in MRS broth and in MRS broth including 0.1%, 0.3% or 0.5% bile salts (Solarbio YZ-1071304) incubated at 37 °C for 6 and 12 h. The inoculums were the precipitated cells of equivoluminal overnight MRS broth cultures (in the early stage of stationary phase) ofL. fermentum9-4 after centrifugation at 4 °C, 8 000 r/min for 5 min and twice washing with 0.9% saline salt. All the experiments were carried out in triplicate.
2.4 Degradation of nucleoside in yogurt fermented by L. fermentum 9-4
The strains with highest nucleoside degradation ability screened by HPLC were cultured in MRS medium at 30 °C for 24 h. Strains of commercially available yogurt: Aiteno probiotic fermented milk (Royal Dairy?of Nanning, China) was inoculated in MRS medium,as a control. After culture, 10 mL culture was centrifuged at 4 °C,4 000 r/min for 10 min to discard medium, the precipitated cells were washed twice with ddH2O and then resuspended in 100 mL pure milk added with 2% sucrose. The mixture was fermented in 37 °C for 12 h, refrigerated at 4 °C for 12 h, and the viable bacteria of the yogurt were counted.
Two milliliters of inosine-guanosine solution (each 1 mmol/L in 50 mmol/L pH 7.5 PBS) was added to 2 mL fermented yogurt, shaking at 37 °C, 160 r/min for 3 h. After centrifuged at 4 °C, 10 000 r/min for 10 min, 300 μL of the supernatant was mixed with 600 μL of 0.6 mol/L perchloric acid, placed on ice for 5 min. Then centrifuged at 4 °C, 10 000 r/min for 1 min, 600 μL of the supernatant was added to 240 μL of 1 mol/L potassium hydroxide solution. After centrifuged at 4 °C, 17 000 r/min for 10 min, the supernatant was filtered (0.22 μm) and analyzed by the same HPLC method as above (see 2.2.4).
2.5 Statistical analysis
Statistical analysis was performed by using three kinds of software, Origin 8.6, GraghPad Prism 6.01 and SPSS 26.0. The significant analysis of variance of test groups was carried out with Duncan test with a con fidence level of 0.05. Data were presented as Mean ± SD values, aP-value less than 0.05 (P< 0.05) was considered as significant.
2.6 Strain and 16S rDNA gene accessions
L. fermentum9-4 has been deposited under the accession number of CCTCC No.: m2019619 in China Center for Type Culture Collection (Wuhan, China). The 16S rDNA of this strain has been submitted to GenBank, and the access number is MT131816.
3. Results
3.1 Screening by selective culture and in vitro purine-metabolizing activity
As shown in Table 1, 10 Shengzhamifen naturally fermented flours were collected from ten old established shops with characteristics of Zhuang nationality in Nanning, China. The pH values of samples distributed between 4.5 (#04 and #06) and 6.8 (#03) with an average ± standard deviation pH value of 5.37 ± 0.87.On the LAB selective MRS medium, the samples showed average viable bacteria counts of (5.83 ± 4.52) × 103CFU/g. Every sample showed very limited kinds of LAB colony morphology on the MRS agar, so only representative colonies were picked out for screening.A total of 35 strains were assayed for their XOD and UOX activities,which are the two key enzymes responsible for the last steps of purine metabolism, catalyzing the successive oxidation of hypoxanthine and xanthine to uric acid, then to water soluble allantoin. As shown in Fig. 1, 6 strains, designated 1-1, 1-2, 1-3, 4-3, 5-1 and 9-4, showed obvious XOD activities, and 4 strains, named 3-2, 4-3, 6-3 and 9-4 showed a significant UOX activities, and their specific activities of XOD and UOX were shown in Table 2. Strains 4-3 and 9-4 showed both the XOD and UOX activities of (0.18 ± 0.02) and (0.38 ± 0.03) U/mg,(0.40 ± 0.06) and (0.29 ± 0.04) U/mg, respectively. All the 8 strains were chosen as candidates for further screening.

Fig. 1 Specific activities of XOD (a) and UOX (b) of 35 candidate strains.The data were presented as the Mean ± SD (n = 3), and were analyzed by using Duncan’s test with a con fidence level of 0.05. The strains sharing the same letter of a-d were not significantly different, while the strains assigned different letters were significantly different with P-value less than 0.05 (P < 0.05).

Table 2XOD and UOX activities of 8 candidate strains.
3.2 Screening and identification of LAB strains by 16S rDNA analysis
To screen out probiotic LAB strains, we carried out 16S rDNA analysis to identify the species of the chosen 8 candidates. As shown in Fig. 2a, genomic DNA with apparent size of around 23.1 kb were extracted from all the 8 strains, and were used as templates to PCR amplify the 16S rDNA genes. All the PCR products appeared the expected approximately 1.5 kb when analyzed by agarose gel electrophoresis (Fig. 2b). After sequencing, the 8 strains were identified by using BLAST to compare their 16S rDNA genes to the GenBank and RDP databases (Table 3). There were 6 strains ofLactobacillus, 1-1, 1-2, 1-3, 4-3, 6-3 and 9-4, a strain ofLeuconostoc citreumand a strain ofBacillus cereus, which were 3-2 and 5-1, respectively. Phylogenetic analysis also showed that 1-2, 1-3, 3-2, 6-3 belonged toL. plantarum, 5-1, 1-1 and 9-4 had the closest relationship withB. cereusEU161996,L. citriumMN135986 andLactobacillus fermentumMF179535, respectively(Fig. 3). All of selected strains are of acid producing bacteria.Since strain 5-1 was a kind of conditional pathogenB. cereus, it was no longer listed as candidate strain, and the other 7 strains remained for next selection.

Fig. 2 Agarose gel electrophoresis of the genome DNA (a) and 16S rDNA PCR products (b) of 8 candidate strains.
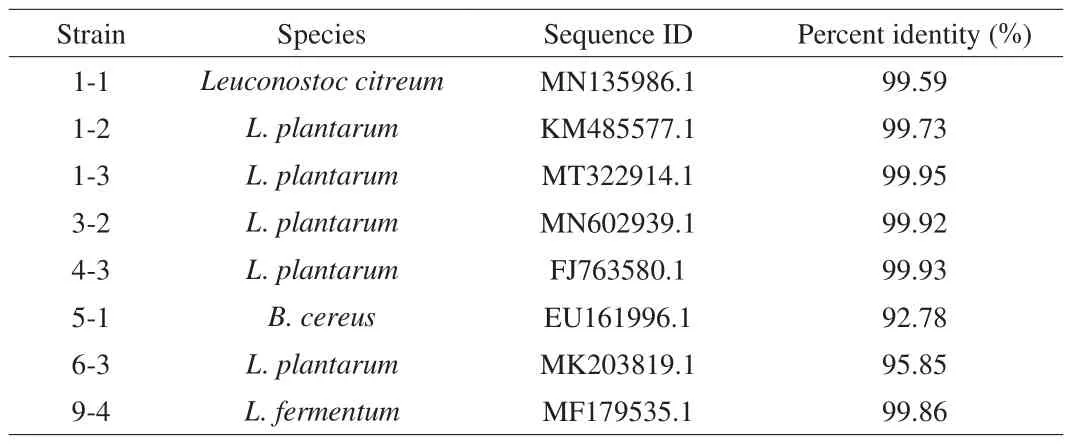
Table 3Strain identification by 16S rDNA analysis.

Fig. 3 Phylogenetic Neighbor-Joining tree of 8 candidate strains based on 16S rDNA sequences. The scale bar 0.05 indicates the nucleotide substitution rate at each site. The phylogenetic tree was constructed using the neighbor-joining method by MEGA-X. The numbers at the nodes are bootstrap con fidence levels (percentage) from 1 000 replicates. The evolutionary distances were computed using the Maximum Composite Likelihood method. Reference sequences were obtained from the GenBank nucleotide sequence database (NCBI).
3.3 Third screening of purine-nucleoside-degrading LAB by HPLC
Seven candidate strains were screened thirdly based on the purine-nucleoside-degrading abilities by HPLC method. As shown in Table 4, all the 7 tested strains could degrade inosine and guanosine,whileL. fermentum9-4 showed the highest degradation rates. The concentration ofL. fermentum9-4 reached 5.02 × 108CFU/mL, whose degradation ratios of inosine and guanosine were (47.74 ± 1.72)%,(58.51 ± 2.38)%, respectively. And degradation speeds of inosine and guanosine were 2.13 × 10-3and 2.78 × 10-3g/(L·min), respectively.As shown in Fig. 4, resting cells ofL. fermentum9-4 decreased dramatically the inosine and guanosine, but almost 100% of consumed inosine was converted to hypoxanthine, while only 2.58% of guanosine was converted to guanine, indicating the ability to degrade inosine into hypoxanthine and assimilate guanosine with a ratio(55.93 ± 3.12)% ofL. fermentum9-4.

Table 4The abilities of candidate LAB strains to degrade inosine and guanosine.

Fig. 4 HPLC analysis of inosine and guanosine degrading abilities of L. fermentum 9-4. (a) Inosine-guanosine solution standards.(b) Inosine-guanosine solution after incubation with L. fermentum 9-4 resting cells for 60 min. (c) Inosine-guanosine solution after incubation with L. fermentum 9-4 resting cells for 120 min.
3.4 Acid and bile salt tolerance of L. fermentum 9-4
To further explore the potential as probiotic, the acid and bile salt tolerances ofL. fermentum9-4 were assayed under the conditions of pH 1.5-3.5 and 0.1%-0.5% bile salt. As shown in Fig. 5a, theL. fermentum9-4 survived very well (> 80%) at pH 2.5 after 3 h treatment, and about 50% could still live after 2 h treatment at pH 1.5. Similarly, more 80% ofL. fermentum9-4 endured the 12 h treatment with 0.1% bile salt under pH 7.5 simulating the intestinal digestion, while only moderate (about 30%) tolerance could keep at the concentration of 0.3% bile salt. The results indicated the good potential probiotic ofL. fermentum9-4 to match the pH of gastric acid and bile salt concentration in intestine, of which the pH fluctuates between 0.9 and 3.5 along with individual’s physical condition and dietary structure, and the concentration of bile salt varies between 0.1% and 0.5% [28].

Fig. 5 Survival of L. fermentum 9-4 after treatment with different pH (a) and different concentrations of bovine bile salt (b).
3.5 Degradation of nucleoside in yogurt fermented by L. fermentum 9-4
To assess the purine degrading ability ofL. fermentum9-4 in foods as probiotics, the yogurt fermented byL. fermentum9-4 was incubated with inosine and guanosine for 3 h. As shown in Fig. 6a,the yogurt 9-4 almost completely consumed the added inosine and guanosine, in comparison to almost no consumption of inosine and guanosine at all in the commercial yogurt (Fig. 6b). The degradation ratios of inosine and guanosine byL. fermentum9-4 yogurt were(98.10 ± 2.58)% and (98.56 ± 3.23)%, respectively.

Fig. 6 HPLC analysis of nucleoside degradation in yogurt fermented by L. fermentum 9-4. (a) Degradation of inosine and guanosine by commercial Aiteno probiotic fermented milk (Royal Dairy? of Nanning, China).(b) Degradation of inosine and guanosine by yogurt fermented by L. fermentum 9-4. Inosine and guanosine were degraded and were not detectable.
4. Discussion
Since 20% uric acid in human body can be reduced by diet control [29], it’s of great significance to reduce purines in food to control serum uric acid level. Probiotics with purine assimilation and degradation ability will help to reduce the absorption of purine nucleoside when ingested with food. Shengzhamifen is a traditional fermented food of Zhuang nationality in China. After a long history of natural domestication, there are a lot of beneficial microorganisms,especially LAB in the naturally fermented rice noodle, making it a good source for probiotic screening. However, the probiotics in Shengzhamifen have not been excavated and applied. In this study,we reported for the first time the isolation of purine-nucleosidedegrading LAB strains from Shengzhamifen. Compared with the previously reported guanosine assimilation rate of 28.61%, which was measured by incubating 15 ODL. fermentumwith 10 mL 2 mmol/L inosine-guanosine mixture for 1 h [21], the assimilation rate ofL. fermentum9-4 to guanosine was 55.93%, to the best of our knowledge,which was also the best one not ever been reported before.
L. fermentum9-4 was screened by combining enzyme activity assay, 16S rDNA molecular identification and HPLC detection,which are usually employed by most of the existingin vitroscreening methods for purine reducing LAB [19]. However, the screening ofL. fermentum9-4 tested the enzymatic activities of XOD and UOX,which consist of the last two steps of purine metabolism and catalyze the purine to water-soluble and easily excreted allantoin. This must be helpful to screen out probiotic strains efficiently degrading purines and lowering uric acid, in considering that purine base can be further metabolized to serum uric acid. The comprehensive method considering the purine nucleoside assimilation, degradation and the metabolic end products proposed here may also benefit the screening of other purine-degrading and uric acid-lowering probiotics.
The degradation of nucleoside is mainly catalyzed by nucleoside hydrolase, which exists in plants and microorganisms, but have not been found in mammals [30].L. fermentum9-4 efficiently assimilated and degraded inosine and guanosine, but only hypoxanthine, the degradation product of inosine was detected during incubation, hardly any guanosine degradation product, guanine, was detected (Fig. 4),indicating the assimilation of guanosine or guanine. The assimilated guanosine and guanine may be finally degraded to allantoin by XOD and UOX in bacterial cells, or guanine may be converted to GMP through purine salvage pathway with the assistance of hypoxanthine guanine phosphoribosyl transferase (HGPRT), and then contribute to the synthesis of DNA and RNA [17,31]. However, the metabolic pathway and mechanism of guanine nucleosides byL. fermentum9-4 need to be further studied.
5. Conclusions
L. fermentum9-4 with purine nucleoside assimilation and degradation ability was firstly screened out by a comprehensive method incorporating enzyme activity assay, 16S rDNA identification and HPLC detection. The resting cells ofL. fermentum9-4 showed the maximum degradation rate of inosine by (47.74 ± 1.72)% and the highest assimilation ratio of guanosine by (55.93 ± 3.12)%, which are improvements over LAB strains characterized previously. Yogurt fermented byL. fermentum9-4 also efficiently assimilated the inosine and guanosine, with respective degradation rates of 98.10% and 98.56% higher than those of the commercial ones. TheL. fermentum9-4 showed excellent survival (> 80%) under the conditions of pH 2.5 and 0.1% bile salt. Although further study of purine reducing mechanism and systemic safety assessment is necessary, our results suggest thatL. fermentum9-4 is a promising candidate as purine-nucleosine-degrading probiotic for the prevention and treatment of hyperuricemia and gout.
conflict of interest
No conflict of interest declared.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (21868003), the Guangxi Natural Science Foundation (2016GXNSFEA380003, 2017GXNSFAA198265,AD18281064) and the Guangxi Science and Technology Major Special Project (AA17204075 and AA17202010-3).
- 食品科學與人類健康(英文)的其它文章
- Hypoglycemic natural products with in vivo activities and their mechanisms: a review
- Bacteroides utilization for dietary polysaccharides and their beneficial effects on gut health
- Capsular polysaccarides of probiotics and their immunomodulatory roles
- Natural compounds may contribute in preventing SARS-CoV-2 infection: a narrative review
- A comprehensive review on the effects of green tea and its components on the immune function
- A review on current and future advancements for commercialized microalgae species

