Alternative Splicing of OsCYL4 Controls Drought Resistance via Regulating Water Loss and Reactive Oxygen Species-Scavenging in Rice
Alternative Splicing ofControls Drought Resistance via Regulating Water Loss and Reactive Oxygen Species-Scavenging in Rice
SUPPLEMENTAL DATA
File S1. Methods
Isolation of OsCYL4b and rice transformation
A full-length cDNA of() isolated from arice variety Zhonghua 11 (ZH11) was obtained from the rice Nipponare cDNA database (http://www.ncgr.ac.cn/ricd/), and the CDS lengths ofandare 804 and 759 bp, respectively. Both constructs for overexpression and suppression ofwere transformed into ZH11 by the-mediated transformation method.The detail information of these primers is shown in Table S2.
Plant growth, treatments, and measurements
The rice plants ZH11 were grown under normal growth conditions, and two-week-old seedlings were treated with abiotic stress to measure the transcript level of. To perform abscisic acid (ABA) treatments of plants at the seedling stage, plantlets that germinated on MS (Murashige and Skoog) medium plates were transferred to MS medium containing 3 μmol/L ABA in autoclaved-visible boxes. The transcript level of target genes was determined using the ricegene () as an internal control in this study, and the detail information of these primers is shown in Table S2. Various treatments and ABA measurement were performed according to the method of Qin et al (2015).
Drought stress was applied to transgenic plants growing in yellow barrel at the seedling stage. Each pot as an independent system was planted with half of the transgenic plants selected by MS medium containing hygromycin and half of the WT plants.After one week, the plants were drought-stressed by depriving water, and the survival rate of the plants was then measured after 7 d of drought treatment and 3 d of recovery, which was significantly different due to the different water contents of soil in each pot.Leaves were cut from plants that had been grown for three weeks under normal conditions to assay water loss in the detached leaves. The leaves were placed on a piece of weighting paper under a light in the greenhouse, immediately weighted after excision, and periodically weighted every hour. The results are shown as a percentage of the fresh weight.
For the analysis of the endogenous ABA level, the four-leaf stage seedlings under normal conditions and drought stress were extracted twice with an extraction buffer. Quantification was performed in an ABI 4000Q-TRAR LC-MS system (Applied Biosystems, USA) with stable isotope-labeled ABA as standards (OlChemIm, Czech Specials) by following the methods described in a previous report (Du et al, 2010).
Subcellular location
The coding region of the genewas amplified to determine the subcellular localization of the OsCYL4b protein. The 35S::OsCYL4b-yellow fluorescence protein (YFP) fusion construct was then produced by inserting the coding region ofinto the vector of pM999 for protoplast transformation and the vector pGreenⅡ0179 for the agroinfiltration method in. Protein OsRac1-C212S (Ono et al, 2001; Wang et al, 2018), CBL1 (Li et al, 2014) and FM4-64 were used as markers or tracker for the cytoplasm and plasma membrane, respectively. The fluorescence signals were captured by a confocal microscopy.
Physiological measurements of guard cells
The leaves of the four-leaf stage plants with the same period of dehydration stress or normal growth were fixed by 2.5% glutaraldehyde, and stomatal images were obtained by a scanning electron microscopy (SEM; JSM-6390LV, JEOL, Japan), as described by Du et al (2010). For the measurement of stomatal conductance, flag leaves of plants at the booting stage were used. The measurements were performed using an LI-6860 porometer (LI-COR, USA).
Reactive oxygen species (ROS) detection
Nitro blue tetrazolium (NBT) staining was performed on rice leaves to detect ROS. Specifically, leaves of three-week-old plants were sampled in the NBT solution. The samples were incubated for 2 h at room temperature, and then maintained in a water bath at 95 oC for 15 min.
Quantitative measurement of H2O2production was performed using an Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Molecular Probes, USA), following the manufacturer’s instructions (Wu et al, 2012). The specific activity of SOD was calculated by dividing the total activity of SOD by the total amount of protein. The total protein content in the enzyme leaching liquor was determined by the method of Bradford (1976).
Bradford M M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding., 72: 248–254.
Du H, Wang N L, Cui F, Li X H, Xiao J H, Xiong L Z. 2010. Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice., 154(3): 1304–1318.
Li J, Long Y, Qi G N, Li J, Xu Z J, Wu W H, Wang Y. 2014. The Os-AKT1 channel is critical for K+uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex., 26(8): 3387–3402.
Ono E, Wong H L, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. 2001. Essential role of the small GTPase Rac in disease resistance of rice., 98(2): 759–764.
Qin Y H, Shen X, Wang N L, Ding X P. 2015. Characterization of a novel cyclase-like gene family involved in controlling stress tolerance in rice., 181: 30–41.
Wang Q, Li Y Y, Ishikawa K, Kosami K I, Uno K, Nagawa S, Tan L, Du J M, Shimamoto K, Kawano Y. 2018. Resistance protein Pit interacts with the GEF OsSPK1 to activate OsRac1 and trigger rice immunity., 115(49): E11551–E11560.
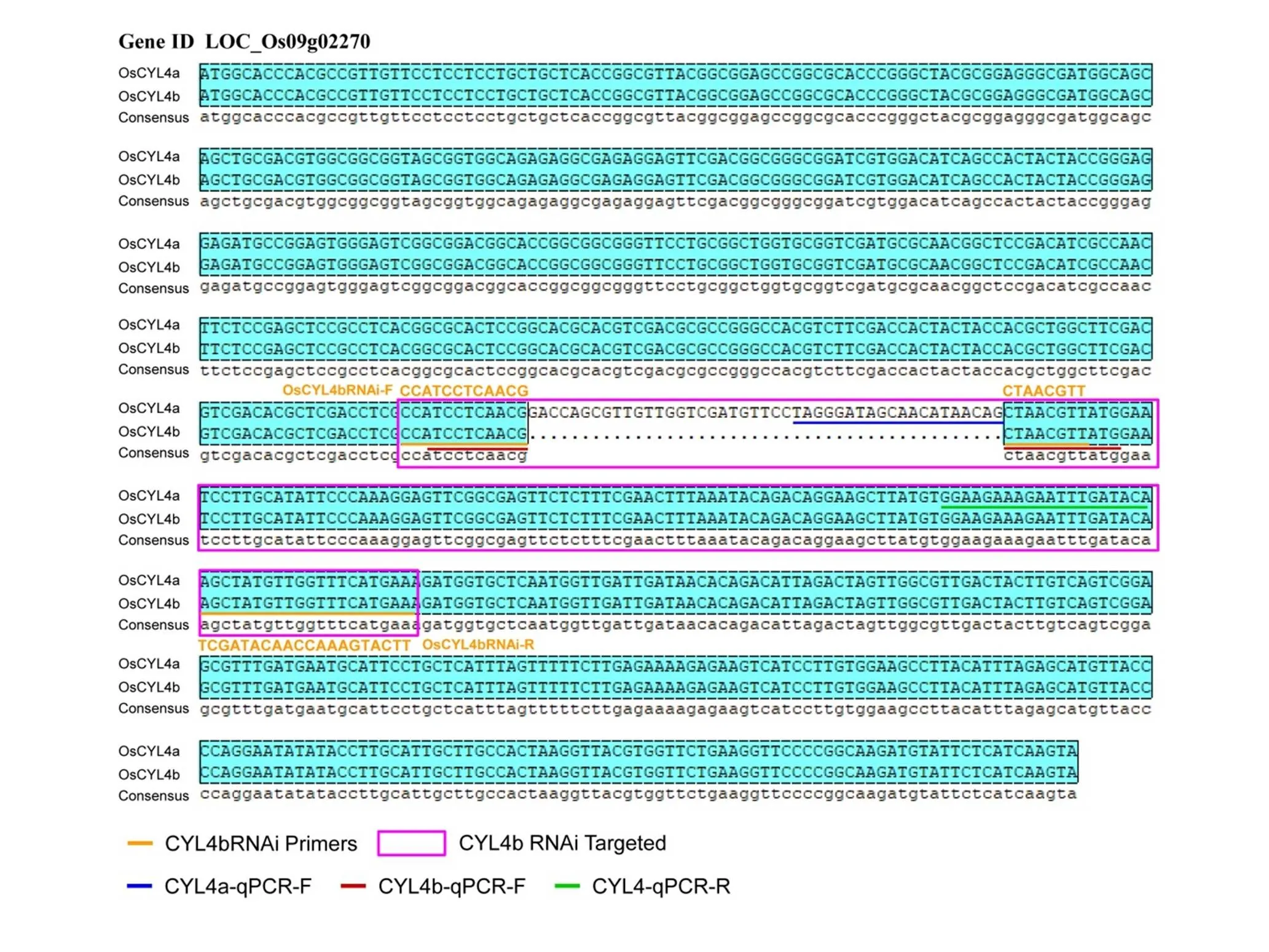
Fig. S1. Sequence alignment with coding sequences ofand
Fig. S2. Expression analysis ofin nine tissues or organs of Zhonghua 11 by qRT-PCR.
1, Root at the tilling stage; 2, Stem at the tilling stage; 3, Sheath; 4, Flag leaf; 5, Leaf; 6, Glume; 7, Young panicle 5; 8, Stamen; 9, Pistil.expression in root at the tillering stage was used as the control among tissues. Ricegene () was used as the endogenous control. Data are Mean ± SE (= 3). *,< 0.05; **,< 0.01.

Fig. S3. Expression analysis ofandresponse to abiotic stresses and abscisic acid (ABA) treatment by qRT-PCR.
Total RNA was extracted from four-week-old leaves. The ricegene () was used as the internal control. *,< 0.05; **,< 0.01.

Fig. S4.Subcellular localization of OsCYL4a and OsCYL4b.
A, Subcellular localization of OsCYL4a-GFP fusion protein in rice protoplasts was analyzed together with plasma membrane tracker FM4-64. Scale bar, 10 μm.
B, Subcellular localization of OsCYL4b-YFP fusion protein inleaf epidermal cells was analyzed together with plasma membrane marker CBL1-CFP. Scale bar, 25 μm.

Fig. S5. Increased tolerance to osmotic stress due tooverexpression.
A and B, Phenotypes of ZH11 andtransgenic plants under normal conditions (A) and mannitol stress (B).
C and D, Seedling height (C) and root length (D) of ZH11 andtransgenic plants under normal conditions and mannitol treatment.
ZH11, Zhonghua 11 (wild type); 4bOE-1 and 4bOE-2,overexpression lines; 4bRNAi-1 and 4bRNAi-2,RNAi lines. Data are Mean ± SE (= 3). *,< 0.05; **,< 0.01.

Fig. S6. Scanning electron microscopy images of three levels of stomatal opening.
Fig. S7. Identification of sensitivity oftransgenic plants to abscisic acid (ABA) treatment.
A, Phenotype of ZH11 andtransgenic plants under normal conditions and ABA treatment.
B, Seedling height and root length of ZH11 andtransgenic plants under normal conditions and ABA treatment.
ZH11, Zhonghua 11 (wild type); 4bOE-1 and 4bOE-3,overexpression lines; 4bRNAi-1 and 4bRNAi-2,RNAi lines. Data are Mean ± SE (= 3). *,< 0.05; **,< 0.01.
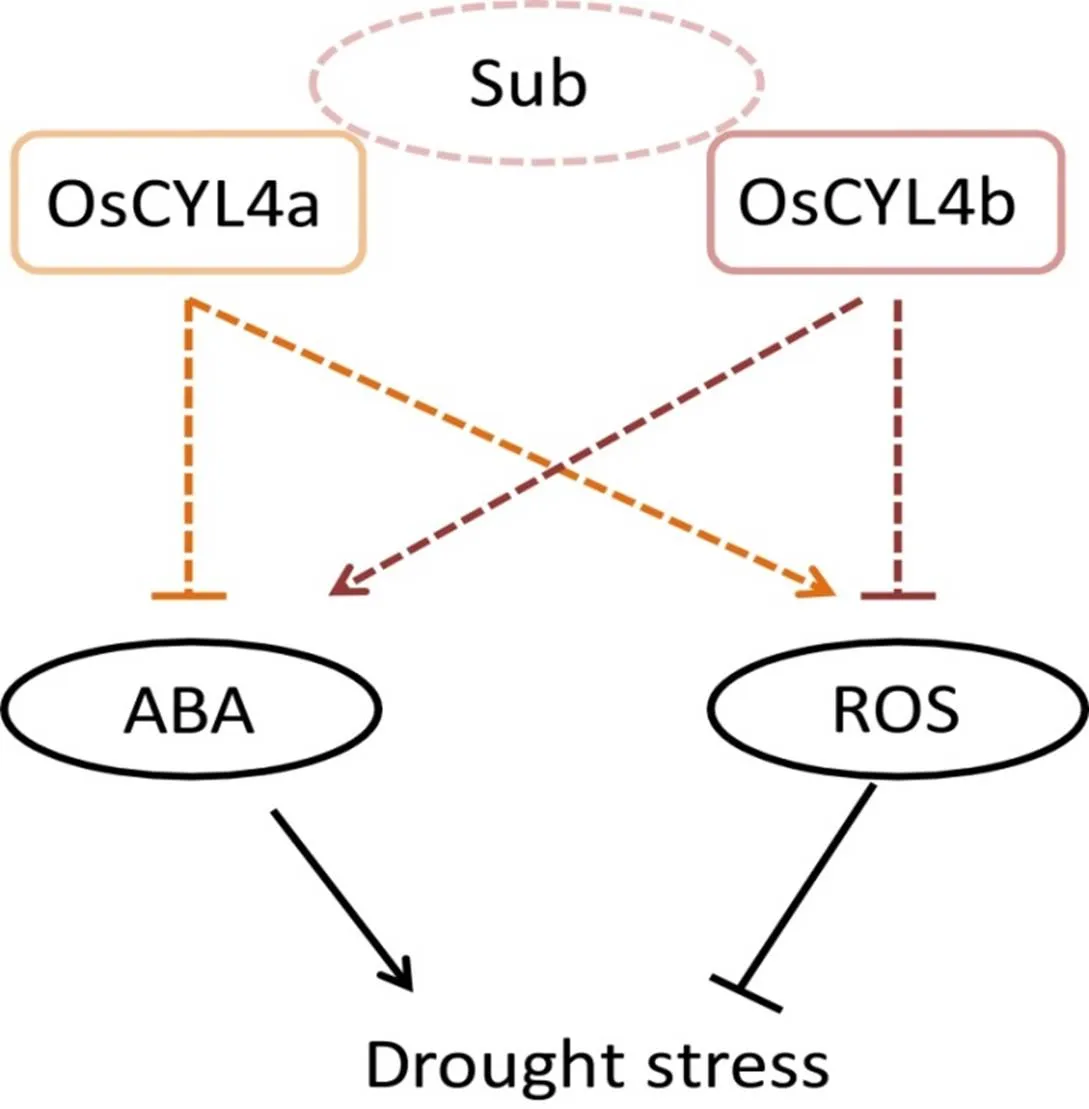
Fig. S8. Functional models including OsCYL4a and OsCYL4b in drought stress.
Sub, Predicted substrates of OsCYL4a like lycopene or polyketide; ABA, Abscisic acid; ROS, Reactive oxygen species.
Table S1.-Elements in 2 kb promoter region ofgene in rice.
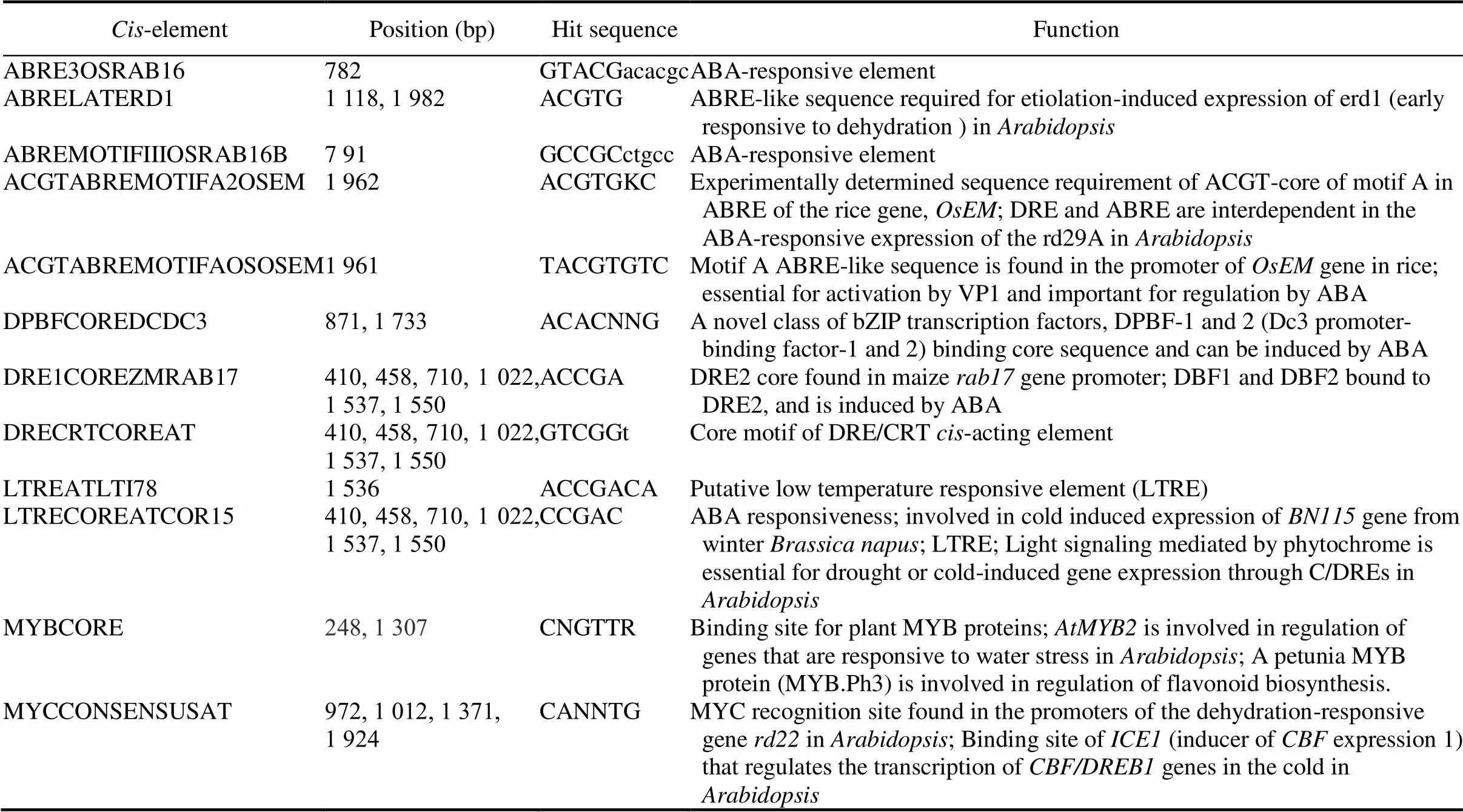
Cis-elementPosition (bp)Hit sequenceFunction ABRE3OSRAB16782GTACGacacgcABA-responsive element ABRELATERD11 118, 1 982ACGTGABRE-like sequence required for etiolation-induced expression of erd1 (early responsive to dehydration ) in Arabidopsis ABREMOTIFIIIOSRAB16B7 91GCCGCctgccABA-responsive element ACGTABREMOTIFA2OSEM 1 962ACGTGKCExperimentally determined sequence requirement of ACGT-core of motif A in ABRE of the rice gene, OsEM; DRE and ABRE are interdependent in the ABA-responsive expression of the rd29A in Arabidopsis ACGTABREMOTIFAOSOSEM1 961TACGTGTCMotif A ABRE-like sequence is found in the promoter of OsEM gene in rice; essential for activation by VP1 and important for regulation by ABA DPBFCOREDCDC3871, 1 733ACACNNGA novel class of bZIP transcription factors, DPBF-1 and 2 (Dc3 promoter- binding factor-1 and 2) binding core sequence and can be induced by ABA DRE1COREZMRAB17410, 458, 710, 1 022, 1 537, 1 550ACCGADRE2 core found in maize rab17 gene promoter; DBF1 and DBF2 bound to DRE2, and is induced by ABA DRECRTCOREAT410, 458, 710, 1 022, 1 537, 1 550GTCGGtCore motif of DRE/CRT cis-acting element LTREATLTI781 536ACCGACAPutative low temperature responsive element (LTRE) LTRECOREATCOR15410, 458, 710, 1 022, 1 537, 1 550CCGACABA responsiveness; involved in cold induced expression of BN115 gene from winter Brassica napus; LTRE; Light signaling mediated by phytochrome is essential for drought or cold-induced gene expression through C/DREs in Arabidopsis MYBCORE248, 1 307CNGTTRBinding site for plant MYB proteins; AtMYB2 is involved in regulation of genes that are responsive to water stress in Arabidopsis; A petunia MYB protein (MYB.Ph3) is involved in regulation of flavonoid biosynthesis. MYCCONSENSUSAT972, 1 012, 1 371, 1 924CANNTGMYC recognition site found in the promoters of the dehydration-responsive gene rd22 in Arabidopsis; Binding site of ICE1 (inducer of CBF expression 1) that regulates the transcription of CBF/DREB1 genes in the cold in Arabidopsis
Table S2. List of all primers used in this study.
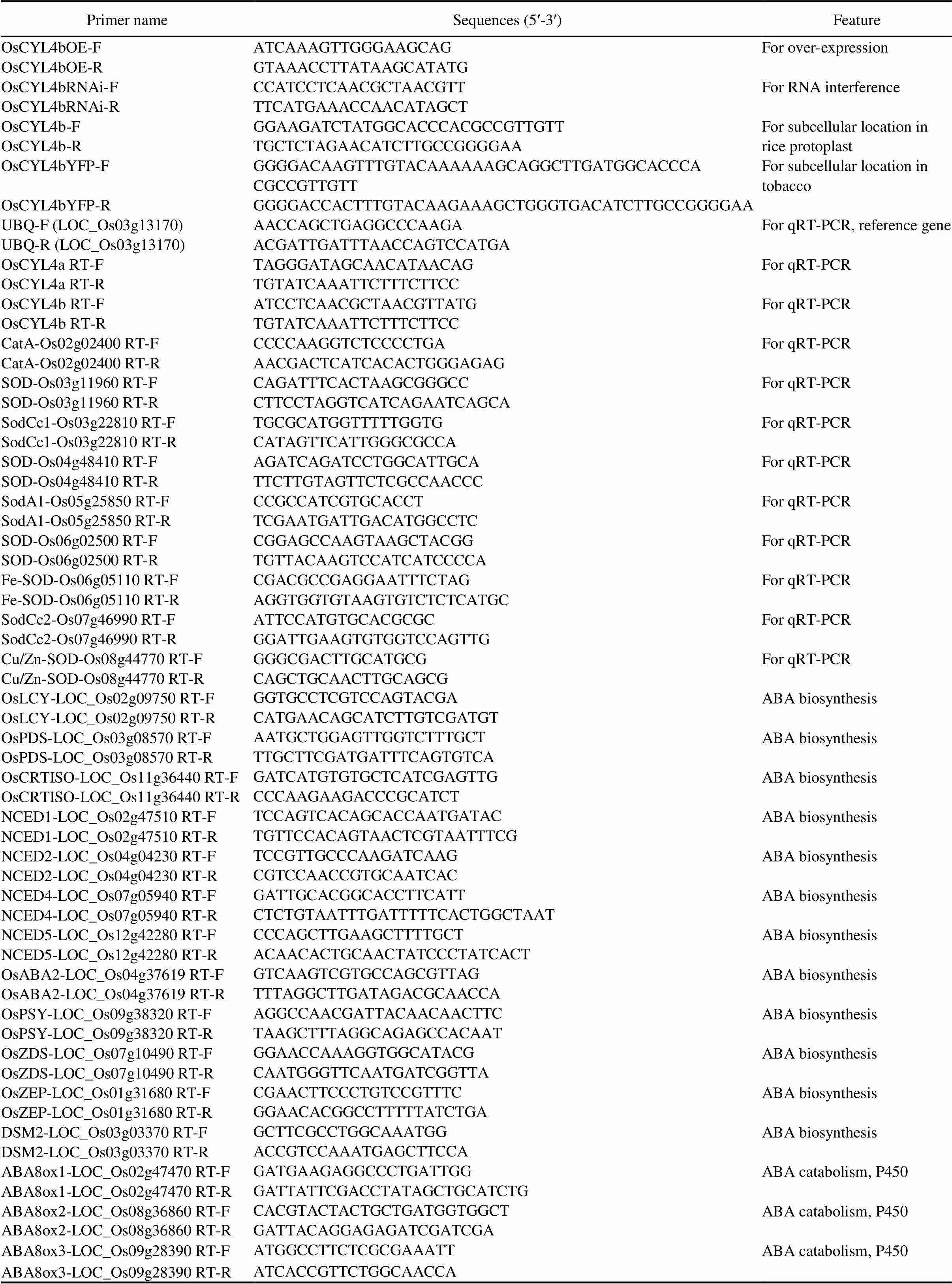
Primer nameSequences (5?-3?)Feature OsCYL4bOE-FATCAAAGTTGGGAAGCAGFor over-expression OsCYL4bOE-RGTAAACCTTATAAGCATATG OsCYL4bRNAi-FCCATCCTCAACGCTAACGTTFor RNA interference OsCYL4bRNAi-RTTCATGAAACCAACATAGCT OsCYL4b-FGGAAGATCTATGGCACCCACGCCGTTGTTFor subcellular location in rice protoplast OsCYL4b-RTGCTCTAGAACATCTTGCCGGGGAA OsCYL4bYFP-FGGGGACAAGTTTGTACAAAAAAGCAGGCTTGATGGCACCCACGCCGTTGTTFor subcellular location in tobacco OsCYL4bYFP-RGGGGACCACTTTGTACAAGAAAGCTGGGTGACATCTTGCCGGGGAA UBQ-F (LOC_Os03g13170)AACCAGCTGAGGCCCAAGAFor qRT-PCR, reference gene UBQ-R (LOC_Os03g13170)ACGATTGATTTAACCAGTCCATGA OsCYL4a RT-FTAGGGATAGCAACATAACAGFor qRT-PCR OsCYL4a RT-RTGTATCAAATTCTTTCTTCC OsCYL4b RT-FATCCTCAACGCTAACGTTATGFor qRT-PCR OsCYL4b RT-RTGTATCAAATTCTTTCTTCC CatA-Os02g02400 RT-FCCCCAAGGTCTCCCCTGAFor qRT-PCR CatA-Os02g02400 RT-RAACGACTCATCACACTGGGAGAG SOD-Os03g11960 RT-FCAGATTTCACTAAGCGGGCCFor qRT-PCR SOD-Os03g11960 RT-RCTTCCTAGGTCATCAGAATCAGCA SodCc1-Os03g22810 RT-FTGCGCATGGTTTTTGGTGFor qRT-PCR SodCc1-Os03g22810 RT-RCATAGTTCATTGGGCGCCA SOD-Os04g48410 RT-FAGATCAGATCCTGGCATTGCAFor qRT-PCR SOD-Os04g48410 RT-RTTCTTGTAGTTCTCGCCAACCC SodA1-Os05g25850 RT-FCCGCCATCGTGCACCTFor qRT-PCR SodA1-Os05g25850 RT-RTCGAATGATTGACATGGCCTC SOD-Os06g02500 RT-FCGGAGCCAAGTAAGCTACGGFor qRT-PCR SOD-Os06g02500 RT-RTGTTACAAGTCCATCATCCCCA Fe-SOD-Os06g05110 RT-FCGACGCCGAGGAATTTCTAGFor qRT-PCR Fe-SOD-Os06g05110 RT-RAGGTGGTGTAAGTGTCTCTCATGC SodCc2-Os07g46990 RT-FATTCCATGTGCACGCGCFor qRT-PCR SodCc2-Os07g46990 RT-RGGATTGAAGTGTGGTCCAGTTG Cu/Zn-SOD-Os08g44770 RT-FGGGCGACTTGCATGCGFor qRT-PCR Cu/Zn-SOD-Os08g44770 RT-RCAGCTGCAACTTGCAGCG OsLCY-LOC_Os02g09750 RT-FGGTGCCTCGTCCAGTACGAABA biosynthesis OsLCY-LOC_Os02g09750 RT-RCATGAACAGCATCTTGTCGATGT OsPDS-LOC_Os03g08570 RT-FAATGCTGGAGTTGGTCTTTGCTABA biosynthesis OsPDS-LOC_Os03g08570 RT-RTTGCTTCGATGATTTCAGTGTCA OsCRTISO-LOC_Os11g36440 RT-FGATCATGTGTGCTCATCGAGTTGABA biosynthesis OsCRTISO-LOC_Os11g36440 RT-RCCCAAGAAGACCCGCATCT NCED1-LOC_Os02g47510 RT-FTCCAGTCACAGCACCAATGATACABA biosynthesis NCED1-LOC_Os02g47510 RT-RTGTTCCACAGTAACTCGTAATTTCG NCED2-LOC_Os04g04230 RT-FTCCGTTGCCCAAGATCAAGABA biosynthesis NCED2-LOC_Os04g04230 RT-RCGTCCAACCGTGCAATCAC NCED4-LOC_Os07g05940 RT-FGATTGCACGGCACCTTCATTABA biosynthesis NCED4-LOC_Os07g05940 RT-RCTCTGTAATTTGATTTTTCACTGGCTAAT NCED5-LOC_Os12g42280 RT-FCCCAGCTTGAAGCTTTTGCTABA biosynthesis NCED5-LOC_Os12g42280 RT-RACAACACTGCAACTATCCCTATCACT OsABA2-LOC_Os04g37619 RT-FGTCAAGTCGTGCCAGCGTTAGABA biosynthesis OsABA2-LOC_Os04g37619 RT-RTTTAGGCTTGATAGACGCAACCA OsPSY-LOC_Os09g38320 RT-FAGGCCAACGATTACAACAACTTCABA biosynthesis OsPSY-LOC_Os09g38320 RT-RTAAGCTTTAGGCAGAGCCACAAT OsZDS-LOC_Os07g10490 RT-FGGAACCAAAGGTGGCATACGABA biosynthesis OsZDS-LOC_Os07g10490 RT-RCAATGGGTTCAATGATCGGTTA OsZEP-LOC_Os01g31680 RT-FCGAACTTCCCTGTCCGTTTCABA biosynthesis OsZEP-LOC_Os01g31680 RT-RGGAACACGGCCTTTTTATCTGA DSM2-LOC_Os03g03370 RT-FGCTTCGCCTGGCAAATGGABA biosynthesis DSM2-LOC_Os03g03370 RT-RACCGTCCAAATGAGCTTCCA ABA8ox1-LOC_Os02g47470 RT-FGATGAAGAGGCCCTGATTGGABA catabolism, P450 ABA8ox1-LOC_Os02g47470 RT-RGATTATTCGACCTATAGCTGCATCTG ABA8ox2-LOC_Os08g36860 RT-FCACGTACTACTGCTGATGGTGGCTABA catabolism, P450 ABA8ox2-LOC_Os08g36860 RT-RGATTACAGGAGAGATCGATCGA ABA8ox3-LOC_Os09g28390 RT-FATGGCCTTCTCGCGAAATTABA catabolism, P450 ABA8ox3-LOC_Os09g28390 RT-RATCACCGTTCTGGCAACCA
- Rice Science的其它文章
- Rice Variety Classification Based on Optimized Near-Infrared Spectral Classification Model
- Effects of Nitrogen-Regulating Gene AreA on Growth,Pathogenicity,and Fumonisin Synthesis of Fusarium proliferatum
- Rice Husk at a Glance:From Agro-Industrial to Modern Applications
- Smart Farming for Sustainable Rice Production:An Insight into Application,Challenge, and Future Prospect
- A β-Carotene Ketolase Gene NfcrtO from Subaerial Cyanobacteria Confers Drought Tolerance in Rice
- OsbZIP01 Affects Plant Growth and Development by Regulating OsSD1 in Rice