Proposal and evaluation of a new norm index-based QSAR model to predict pEC50and pCC50activities of HEPT derivatives☆
Kanwal Shahid,Qiang Wang,*,Qingzhu Jia,Lei Li,Xue Cui,Shuqian Xia,Peisheng Ma
1School of Chemical Engineering and Material Science,Tianjin University of Science and Technology,13St.TEDA,Tianjin 300457,China
2School of Marine and Environment Science,Tianjin University of Science and Technology,13St.TEDA,Tianjin 300457,China
3School of Chemical Engineering and Technology,Tianjin University,Tianjin 300072,China
1.Introduction
Being as a global epidemic, acquired immunodeficiency syndrome(AIDS) is considered as one of the worst diseases ever known to mankind[1]. AIDS is a collection of symptoms and infections resulting from the specific damage to the immune system caused by the human immunodeficiency virus (HIV) [2,3], which is a member of a retroviruses group. Retroviruses contain special single stranded RNAs (genetic materials of a virus) which target the host cells; once in the host cells, they use their own enzymes called as “Reverse transcriptase” to convert the virus RNA to proviral DNA. In the HIV life cycle, three enzymes are essential for replication of this virus inside the host cells, reverse transcriptase (RT), protease(PR) and integrase (IN). Theoretically, an anti-HIV agent may exert its activity by inhibiting a variety of steps in the life cycle of the virus;which is why, this stage of virus life cycle is considered to be one of the prime and promising targets for the development of anti-HIV drugs [4–6].
In order to search for anti-HIV drugs with fewer side effects and high efficacy, modeling the biological activity to propose new candidate molecules is an important approach. So, over the last few years, the quantitative structure–activity relationship (QSAR) studies have been widely carried out for different series of HIV-1 inhibitors, such as HIV-1 RT inhibitors(RTIs) [7–9], HIV-1 IN inhibitors [10–12] and HIV-1 protease inhibitors [13].
Certainly,a large number of compounds have already been synthesized to target various HIV-1-RT active sites,for example,HEPT derivatives(1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine)are some of those several proved potent HIV-NNRTIs(non-nucleoside inhibitors).Generally,in order to evaluate the performance of anti-HIV drugs,two important values,the required effective concentration to achieve 50%protection of MT-4 cells against the cytopathic effect of virus(EC50,expressed as–lgEC50,pEC50)and the required cytotoxic concentration to reduce visibility of 50%mock infected cell(CC50,expressed as–lgCC50,pCC50)were widely utilized,and some QSAR investigates have been successfully performed in this field[14–16].For instance,Bazoui et al. [17] employed multiple linear regression (MLR) and artificial neural network (ANN) approaches to develop two models for predicting pEC50of 95 HEPT derivatives, and their models were very satisfying with good statistical values (the squared correction coefficient R2of 0.83 and 0.85 in MLR and ANN model,respectively).In Chen et al.'s work[18],two nonlinear models based on Comparative Molecular Field Analysis(CoMFA)and Comparative Molecular Similarity Index Analysis(CoMSIA)were proposed for pEC50prediction of the data set of 88 compounds with R2of 0.926 and 0.954,respectively.Also,in Guo et al.'s investigation[19],by using the supervised stochastic resonance(SSR)approach,a QSAR model(R2=0.8858)was developed for predicting pEC50of 80 HEPT derivatives.
Recently,based on the molecule's distance matrix and atom characters matrix,our group has proposed some norm index-based models which were successfully utilized for predicting different properties of compounds including the narcotic pollutants' aquatic toxicity[20],the pharmacological and toxicological activity of heterocyclic compounds[21],and high affinity 5-HT1Areceptor ligands of arylpiperazine derivatives[22].Our previous work suggested that this new approach might be capable to be further utilized in a large field.
Therefore,this research work was carried out to satisfy two goals:(1)to propose a new norm index,(2)to develop a more accurate and stable structure–activity relationship model for biological activity prediction of HIV-1 inhibitors HEPT derivatives.
2.Methods
2.1.Data Sets
This research work was carried out to predict the pEC50(134 compounds)and pCC50(39 compounds)activities of HEPT derivatives,and their general structure is shown in Fig.1;the observed and predicted values of pEC50and pCC50activities of these compounds are presented in Table S1 and Table S2(provided as Supporting Information),respectively[13,23–26].For both pEC50and pCC50prediction,the training set and test set were divided randomly as same as those of the reference work[13].

Fig.1.The general structure of HEPT derivatives.
The molecular structures were drawn using the free version of Hyperchem(http://www.hyper.com/)[27].The molecules were then pre-optimized using the molecular mechanics force field(MM+)calculations of the software. Energy minimization of the molecules was obtained by using ab-initio method. Among which, the charge distribution and the molecules' geometries were optimized by using ab initio methods at STO-3G level and a gradient norm limit of 4.184×109kJ·m?1was set for the purpose.
2.2.Model Construction
In order to illustrate the atom distribution and constituent of a molecule clearly and quantitatively,some step distance matrixes and a property matrix of a molecule have been proposed and used in our QSAR approach.Firstly,the step distance matrices of HEPT derivatives' structures were generated based on their chemical graphs.Here,the step distance matrix consisted of the adjacent step distance matrix,the interval step distance matrix and the interval jump step distance matrix shown as Eq.(1).Then,a property matrix including various atomic characters such as atomic weight,van der Waals radius,electronegativity and atom charge,were specially defined in order to improve the predictive effect of this method.In addition,it was obvious that information concerning atom/heteroatom connectivity patterns and hybrid electronic information in the molecule could be encoded by these atomic characters.
The step distance matrices and the property matrix used in this research were shown as follows:

where eiis atom i's electronegativity.
Based on Eqs.(1)and(2),the extended distance matrices MD(including 10 matrices)were further de fined,then,some norm indexes of the above 10 MD matrices were also proposed and listed as in Table 1.In this work,three kinds of norm indexes are specially de fined:the norm(MD,1)means the largest column sum of matrix MD,the norm(MD,2)stands for the largest singular value of matrix MD,and the norm(MD,fro)is the Frobenius-norm of matrix MD.

Table 1 Norm indexes of extended distance matrices MD and parameters of Eq.(3)
According to these norm indexes,a multiple regression QSAR mathematical model was developed and expressed as Eq.(3):

where,lg(1/C)stands for the two biological activities of pEC50or pCC50,b0is the constant,MDiis the descriptor and biis the corresponding regression coefficient of this MLR model.The individual values of all variables are also listed as in Table 1.During our modeling work,two arithmetic linear methods,MLR(multiple linear regression)and PLS(partial least-squares)approaches were used and surprising similar results have been obtained by using the two different regression methods.Therefore,all the prediction results provided in this work were just based on the MLR approach.
2.3.Model Validation
The quality of this model was determined by the statistical values of regression model,leave-one-out cross-validation(LOO-CV)and Y-randomization test.

Table 2 Statistical results for prediction of pEC50and pCC50for HEPT derivatives based on this model and the references' models
2.4.Applicability Domain(AD)
The applicability domain (AD) determines the predictive power of the QSAR model.Verification of applicability domain is essential,especially if the model is to be used to screen new compounds.In this research work,the AD of the predictive model was verified by the leverage approach using a Williams graph[28],where the leverage values(h)were plotted against the training set's standard residual values.In this plot,the AD is established inside a calculated area(leverage threshold,defined as h*=3((N+1)/n))within±3 standard deviations.The compounds outside this area(i.e.the leverage of the compound is greater than leverage threshold,i.e.h>h*)are treated as outliers.The individual leverage threshold values are given with the relative plots[28,29].
3.Results and Discussion
3.1.Prediction Results of pEC50and pCC50
The pEC50and pCC50prediction results of this model were listed in Table S1 and Table S2.And statistical metrics for the predictive model R2and ARD were summarized in Table 2.The predicted versus experimental values scatter diagram for this regression were presented in Figs.2 and 3.Figs.4 and 5 showed the plot of residuals versus experimental values.For convenient utilization of our model,the pEC50and pCC50prediction process were described in detail as Appendix A.
Results in Fig.2 indicate that the predicted pEC50agrees well with the experimental results for 134 HEPT derivatives. Statistical metrics described that our model could give satisfactory prediction results of pEC50withof 0.774,respectively.Fig.4 showed that prediction residuals of our model for pEC50were between?1 and 1 for the most of HEPT derivatives except for three compounds.Also,by using Eq.(3),the pCC50values could be predicted well as shown in Fig.3 and our satisfactory prediction results could be testified by theand the lower prediction residuals as showed in Table 2 and Fig.5.

Fig.2.The predicted vs.experimental pEC50values for 134 HEPT derivatives.

Fig.3.The predicted vs.experimental pCC50values for 39 HEPT derivatives.

Fig.5.Plot of the residual vs.experimental pCC50from this model.
Also,in order to compare with other models,some reference methods[14,17,19,30,31]and their regression statistical results for pEC50and pCC50prediction were listed in Table 2.In case of pEC50,Table 2 showed that the predictive ability of our method(R2of 0.847)was better than MLR-based linear modeling method(R2of 0.83).While methods based on ANN,NN and SSR approaches[14,17,19]could give better prediction results with R2of 0.977,0.85 and 0.886,respectively.However,it was obvious that all the dataset considered in their works[14,17,19]had not been separated into training and test sets,which might limit the applicability of these methods to some degree;accordingly,the predictive capability of MLR-based linear modeling method was not good enough(Q2of 0.70).While for pCC50prediction,our model outperformed(R2of 0.815)the reference methods(R2=0.78–0.81)whether it was a linear or non-linear model.On the whole,the methodologies used in the other studies were very different;it is certain that each method had its merit,and these methods might have the optional application fields for pEC50and pCC50prediction for special HEPT derivatives with accuracy.What's more,it should be pointed that our method was a linear model and could be expressed concretely as a formula,which could be further used by others conveniently if comparing with these nonlinear methods(ANN and NN).
3.2.Leave-one-out Cross-validation
Being as a model validation technique,cross-validation approach is mainly utilized to estimate how accurately a predictive model will perform in practice.And the objective of cross-validation is to de fine a dataset to “test”the model in the training phase,in order to limit problems like over fitting during modeling work. For instance, the leave-one out cross-validation(LOO-CV)approach is a powerful general technique and widely applied for the model evaluation. Generally, during the LOOCV process,only one sample is used as the test set,and the remaining(N-1)samples are becoming the training set if the original dataset is of N samples.Then,N new models would be developed,accordingly,N statistical values of Q2(the squared correlation coefficient of LOO-CV)would be obtained.Lastly,the average of N statistical values of Q2would be set as the final LOO-CV validation result.

where,Yobs,Ypredand Y stand for observed,predicted and the mean observed activities,respectively.
The predictive ability of this model is validated by LOO-CV as shown in Table 2.And distributions of the relative derivation(RD)by LOO-CV and this model for pEC50and pCC50were presented as Figs.6 and 7.The higher Q2of0.787and0.846values for pEC50 and pCC50prediction obtained from LOO-CV suggested the reliability of our model.Also,from Figs.6 and 7,it was obvious that RD distributions for both LOO-CV prediction and our model prediction were very similar,which further demonstrated the stability of our norm-index-based model for prediction pEC50and pCC50for these HEPT derivatives.

Fig.6.Distributions of the relative derivation(RD)by leave-one-out cross-validation and this model for pEC50.

Fig.7.Distributions of the relative derivation(RD)by leave-one-out cross-validation and this model for pCC50.
3.3.Y-randomization Test
Usually,Y-randomization test technique was performed in order to avoid the possibility of chance correlation for the modeling work;and also,this approach was widely utilized to evaluate the robustness of QSAR model.The dependent variable vector(training set compounds)is shuffled randomly to create a new QSAR model using the independent variable matrix.Generally,the lower R2and Q2values of bothprediction and our model prediction were very similar,which further demonstrated the stability
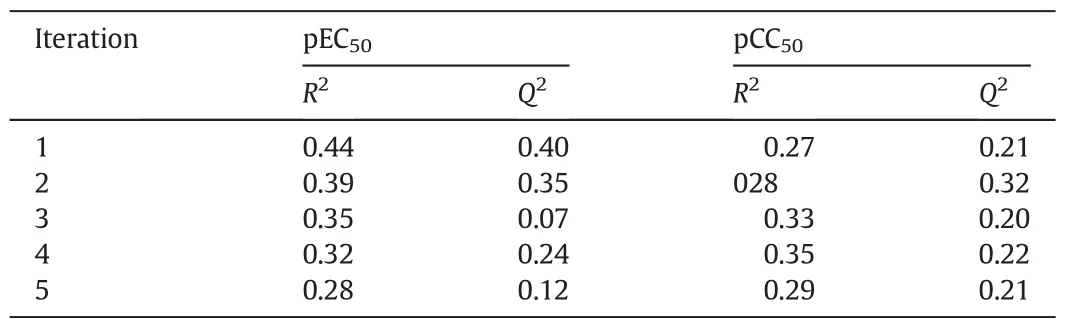
Table 3The Y-randomization test results to validate the model robustness to predict pEC50and pCC50
In this work, five random shuffles of the y vector were carried out at 95%confidence level for each QSAR dataset and results were listed in Table 3.Results shown in Table 3 suggested that values of R2and Q2of these new random models were Significant lower than those of our original model both for pEC50and pCC50.Accordingly,our QSAR model is robust and there was not chance correlation during our modeling work.
3.4.Applicability Domain(AD)
The applicability domain of this proposed QSAR model forp EC50and pCC50was verified by the Williams graph and the plot of the diagonal values of the hat matrix(H)versus standardized residuals was shown as Figs.8 and 9.Results of Fig.8 described that most of the training set substances and test set were included in the AD of this model;only two training compounds(the compound Nos.of 61 and 100)and one test compound(the compound No.of 131)were identified and verified as structural outliers for pEC50prediction.As for pCC50prediction,all the 39 compounds were distributed in the AD of this model.Consequently,it could be deduced that this developed QSAR models could cover a large response and structural applicability domain both for pEC50and pCC50prediction of HEPT derivatives.
4.Conclusions
In this study, based on the norm indexes proposed by authors, a new QSAR model was developed for predicting the pEC50and pCC50activities of more than 150 HEPT derivatives(1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine).Results indicated that this new model could provide satisfactory results for prediction of pEC50and pCC50with theComparison results with reference methods demonstrated that this new method could result in improvements for predicting pEC50and pCC50of anti-HIV HEPT derivatives.Leave-one-out cross validation and Y-randomization test results suggested the reliability and stability of our model,and this model might be applied in a large response and structural domain by verified applicability domain.In summary,these validation results prove that this model might be potent and could be further used to study other activities of related HEPT derivatives.


Fig.8.Applicability domain of our model for pEC50prediction of 134 HEPT derivatives.

Fig.9.Applicability domain of our model for pCC50prediction of 39 HEPT derivatives.

Appendix A
Prediction for pEC50andpCC50of the first compound in Table S1 and Table S2:
The structure of this compound is as follows:

Firstly,four step distance matrices and a property matrix Meof this compound were generated based on it's chemical graph shown as Eqs.(1)and(2).Then,based on Eqs.(1)and(2),the extended distance matrices then,some norm indexes(including the norm(MD,1),the norm(MD,2)and the norm(MD,fro))of the above 10 MD matrices were calculated and listed in Table 4.

Table 4Norm indexes values of the 10 extended distance matrices MD for the first compound in Table S1 and Table S2
Based on parameters shown in Tables 1 and 4,the pEC50and pCC50of this compound was predicted by Eq.(3):

The calculated pCC50result is 3.64,while the experimental pCC50is 3.52.
Appendix B.Supplementary data
The observed and predicted pEC50values of 134 HEPT derivatives are listed in Table S1,the observed and predicted pCC50values of 39 HEPT derivatives are listed in Table S2.Supplementary data to this article can be found online at doi:http://dx.doi.org/10.1016/j.cjche.2016.04.010.
[1]http://www.unaids.org/en/dataanalysis.
[2]M.Baba,H.Tanaka,E.De Clercq,R.Pauwels,J.Balzarini,D.Schols,H.Nakashima,C.F.Perno,R.Walker,T.Miyasaka,Highly specific inhibition of human immunodeficiency virus type 1 by a novel 6-substituted acyclouridine derivative,Biochem.Biophys.Res.Commun.165(1989)1375–1381.
[3]World Health Organization,J.U.N.P.o.,UNICEF,Global HIV/AIDS response:Epidemic update and health sector progress towards universal access:Progress report 2011,World Health Organization,2011.
[4]T.Miyasaka,H.Tanaka,M.Baba,H.Hayakawa,R.T.Walker,J.Balzarini,E.De Clercq,A novel lead for specific anti-HIV-1 agents:1-[(2-Hydroxyethoxy)methyl]-6-(phenylthio)thymine,J.Med.Chem.32(1989)2507–2509.
[5]C.M.Bailey,T.J.Sullivan,P.Iyidogan,J.Tirado-Rives,R.Chung,J.Ruiz-Caro,E.Mohamed,W.Jorgensen,R.Hunter,K.S.Anderson,Bifunctional inhibition of human immunode fi ciency virus type 1 reverse transcriptase:Mechanism and proof-of-concept as a novel therapeutic design strategy,J.Med.Chem.56(2013)3959–3968.
[6]K.M.Frey,D.E.Puleo,K.A.Spasov,M.Bollini,W.L.Jorgensen,K.S.Anderson,Structure-based evaluation of non-nucleoside inhibitors with improved potency and solubility that target HIV reverse transcriptase variants,J.Med.Chem.58(2015)2737–2745.
[7]L.He,P.C.Jurs,Assessing the reliability of a QSAR model's predictions,J.Mol.Graph.Model.23(2005)503–523.
[8]A.Golbraikh,A.Tropsha,Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection,J.Comput.Aided Mol.Des.16(2002)357–369.
[9]A.Golbraikh,A.Tropsha,Beware of q2!J.Mol.Graph.Model.20(2002)269–276.
[10]A.Golbraikh,M.Shen,Z.Xiao,Y.-D.Xiao,K.-H.Lee,A.Tropsha,Rational selection of training and test sets for the development of validated QSAR models,J.Comput.Aided Mol.Des.17(2003)241–253.
[11]S.Raic-Malic,D.Svedruzic,T.Gazivoda,A.Marunovic,A.Hergold-Brundic,A.Nagl,J.Balzarini,E.De Clercq,M.Mintas,Synthesis and antitumor activities of novel pyrimidine derivatives of 2,3-O,O-dibenzyl-6-deoxy-L-ascorbic acid and 4,5-didehydro-5,6-dideoxy-L-ascorbic acid,J.Med.Chem.43(2000)4806–4811.
[12]L.Eriksson,J.Jaworska,A.P.Worth,M.T.Cronin,R.M.McDowell,P.Gramatica,Methods for reliability and uncertainty assessment and for applicability evaluations of classi fi cation-and regression-based QSARs,Environ.Health Perspect.111(2003)1361.
[13]R.Garg,S.P.Gupta,H.Gao,M.S.Babu,A.K.Debnath,C.Hansch,Comparative quantitative structure–activity relationship studies on anti-HIV drugs,Chem.Rev.99(1999)3525–3602.
[14]L.Douali,D.Villemin,D.Cherqaoui,Neural networks:Accurate nonlinear QSAR model for HEPT derivatives,J.Chem.Inf.Comput.Sci.43(2003)1200–1207.
[15]V.P.Solov'ev,A.Varnek,Anti-HIV activity of HEPT,TIBO,and cyclic urea derivatives:Structure–property studies,focused combinatorial library generation,and hits selection using substructural molecular fragments method,J.Chem.Inf.Comput.Sci.43(2003)1703–1719.
[16]Y.Akhlaghi,M.Kompany-Zareh,Application of radial basis function networks and successive projections algorithm in a QSAR study of anti-HIV activity for a large group of HEPT derivatives,J.Chemom.20(2006)1–12.
[17]H.Bazoui,M.Zahouily,S.Boulajaaj,S.Sebti,D.Zakarya,QSAR for anti-HIV activity of HEPT derivatives,SAR QSAR Environ.Res.13(2002)567–577.
[18]H.F.Chen,X.J.Yao,Q.Li,S.G.Yuan,A.Panaye,J.P.Doucet,B.T.Fan,Comparativestudy of non-nucleoside inhibitors with HIV-1 reverse transcriptase based on 3D-QSAR and docking,SAR QSAR Environ.Res.14(2003)455–474.
[19]W.Guo,X.Hu,N.Chu,C.Yin,Quantitative structure–activity relationship studies on HEPTs by supervised stochastic resonance,Bioorg.Med.Chem.Lett.16(2006)2855–2859.
[20]Q.Wang,Q.Jia,L.Yan,S.Xia,P.Ma,Quantitative structure–toxicity relationship of the aquatic toxicity for various narcotic pollutants using the norm indexes,Chemosphere 108(2014)383–387.
[21]Z.C.Zhu,Q.Wang,Q.Z.Jia,S.Q.Xia,P.S.Ma,Structure–property relationship for the pharmacological and toxicological activity of heterocyclic compounds,Acta Phys.-Chim.Sin.30(2014)1086–1090.
[22]Q.Jia,X.Cui,L.Li,Q.Wang,Y.Liu,S.Xia,P.Ma,A quantitative structure–activity relationship for high affinity 5-HT1A receptor ligands based on norm indexes,J.Phys.Chem.B 119(2015)15561–15567.
[23]H.Tanaka,H.Takashima,M.Ubasawa,K.Sekiya,I.Nitta,M.Baba,S.Shigeta,R.T.Walker,E.De Clercq,T.Miyasaka,Synthesis and antiviral activity of deoxy analogs of I-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine(HEPT)as potent and selective anti-HIV-1 agents,J.Med.Chem.35(1992)4713–4719.
[24]M.Baba,S.Shigeta,H.Tanaka,T.Miyasaka,M.Ubasawa,K.Umezu,R.T.Walker,R.Pauwels,E.De Clercq,Highly potent and selective inhibition of HIV-1 replication by 6-phenylthiouracil derivatives,Antivir.Res.17(1992)245–264.
[25](a)T.Miyasaka,H.Tanaka,M.Baba,H.Hayakawa,R.T.Walker,J.Balzarini,E.De Clercq,A novel lead for speci fi c anti-HIV-1 agents:1-[(2-Hydroxyet hoxy)methyl]-6-(phenylthio)thymine,J.Med.Chem.32(1989)2507–2509;
(b)M.Baba,H.Tanaka,E.DeClercq,R.Pauwels,J.Balzarini,D.Schols,H.Nakashima,C.F.Perno,R.T.Walker,T.Miyasaka,Highly specific inhibition of human immunodeficiency firus type 1 by a novel substituted acyclouridine derivative,Biochem.Biophys.Res.Commun.165(1989)1375–1381;
(c)M. Tanaka, M. Baba, M. Ubasawa, H. Takashima, K. Sekiya, I. Nitta, S. Shigeta, R.T.Walker, E. De Clercq, T.Miyasaka, Synthesis and anti-HIV activity of 2-,3-, and 4-substituted analogues of 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine(HEPT), J. Med. Chem. 34 (1991) 1394–1399;
(d)H.Tanaka,M.Baba,S.Saito,T.Miyasaka,H.Takashima,K.Sekiya,M.Ubasawa,I.Nitta,R.T.Walker,H.Nakashima,E.De Clercq,Specificanti-HIV-1“Acyclonucleosides”which cannot be phosphorylated:Synthesis of some deoxy analogues of 1-[(2-Hydroxyethoxy)methyll-6-(phenylthio)thymine,J.Med.Chem.34(1991)1508–1511;
(e)H.Tanaka,M.Baba,H.Hayakawa,K.Haraguchi,T.Miyasaka,M.Ubasawa,H.Takashima,K.Sekiya,I.Nitta,R.T.Walker,E.De Clercq,Lithiation of uracilnucleosides and its application to the synthesis of a new class of anti-HIV-1 acyclonucleosides,Nucleosides Nucleotides 10(1991)397–400.
[26]J.M.Luco,F.H.Ferretti,QSAR based on multiple linear regression and PLS methods for the anti-HIV activity of a large group of HEPT derivatives,J.Chem.Inf.Comput.Sci.37(1997)392–401.
[27]Hyperchem.7.0.Hypercube,Inc.,http://www.hyper.com 2001.
[28]P.Gramatica,Principles of QSAR models validation:Internal and external,QSAR Comb.Sci.26(2007)694–701.
[29]P.Gramatica,E.Giani,E.Papa,Statistical external validation and consensus modeling:A QSPR case study for Koc prediction,J.Mol.Graph.Model.25(2007)755–766.
[30]H.Bazoui,M.Zahouily,S.Sebti,S.Boulajaaj,D.Zakarya,Structure–cytotoxicity relationships for a series of HEPT derivatives,J.Mol.Model.8(2002)1–7.
[31]V.K.Agrawal,J.Singh,K.Mishra,P.V.Khadikar,QSAR study on cytotoxic activities of a series of HEPT analogues,Lett.Drug Des.Discovery 3(2006)129–137.
[32]C.Rücker,G.Rücker,M.Meringer,Y-randomization and its variants in QSPR/QSAR,J.Chem.Inf.Model.47(2007)2345–2357.
[33]A.Tropsha,P.Gramatica,V.K.Gombar,The importance of being earnest:Validation is the absolute essential for successful application and interpretation of QSPR models,QSAR Comb.Sci.22(2003)69–77.
 Chinese Journal of Chemical Engineering2016年10期
Chinese Journal of Chemical Engineering2016年10期
- Chinese Journal of Chemical Engineering的其它文章
- CFD modeling of a headbox with injecting dilution water in a central step diffusion tube☆
- Interactions between two in-line drops rising in pure glycerin☆
- Hydrodynamics of three-phase fluidization of homogeneous ternary mixture in a conical conduit—Experimental and statistical analysis
- Adsorption of Hg(II)from aqueous solution using thiourea functionalized chelating fiber☆
- Nickel(II)removal from water using silica-based hybrid adsorbents:Fabrication and adsorption kinetics☆
- Reactive dividing wall column for hydrolysis of methyl acetate:Design and control☆
