Molecular Phylogeny and Evolution of Two Rhacophorus Species Endemic to Mainland Japan
Masafumi MATSUI, Yasuhiro KAWAHARA, Kanto NISHIKAWA, Seiji IKEDA,Koshiro ETO and Yusuke MIZUNO
1 Graduate School of Human and Environmental Studies, Kyoto University, Yoshida Nihonmatsu-cho, Sakyo-ku, Kyoto 606-8501, Japan
2 Hagi City Museum, Horiuchi 355, Hagi, Yamaguchi 758-0057, Japan
3 Hiroshima University Museum, Kagamiyama 1-1-1, Higashi Hiroshima City, Hiroshima 739-8524, Japan
4 Kitakyushu Museum of Natural History and Human History, Higashida 2-4-1, Yahatahigashi-ku, Kitakyushu, Fukuoka 805-0071, Japan
5 Takahari 4-1002, Meito-ku, Nagoya 465-0061, Japan
Abstract We conducted molecular phylogenetic analyses of Japanese Rhacophorus species, especially of R. schlegelii and R. arboreus from the mainland, based on samples encompassing their known distribution ranges, and discussed about evolutionary history of Rhacophorus species within Japan. The common ancestor of Japanese Rhacophorus,except for R. owstoni from southern Ryukyus, was estimated to have diverged from a lineage occurring mainly in China about 7 MYBP. Both R. schlegelii and R. arboreus are genetically largely divergent between regions of eastern and western Japan, and this seems to have been promoted mainly by retreat to refugia. Retreats of the two species to different refugia sometimes in the past seem to have led restricted distribution of R. schlegelii in eastern and R. arboreus in western Japan, and brought their intraspecific variation patterns in morphology and breeding habit.
Keywords glacial period, mitochondrial DNA differentiation, refugia, Rhacophorus arboreus, Rhacophorus schlegelii
1. Introduction
Some amphibians like the newt genus Cynops and the frog family Rhacophoridae have Japanese mainland as their northern limit of distribution. It is suggested that some species of such groups contain populations that were affected by climate changes in the Quaternary glacier and interglacial periods (e.g., Cynops pyrrhogaster[Tominaga et al., 2013]; Buergeria buergeri [Nishizawa et al., 2013]). A rhacophorid genus Rhacophorus occurs from Southeast to East Asia and contains about 92 species(Frost, 2019). In Japan, R. owstoni, R. viridis, and R.amamiensis occur in the Ryukyu Archipelago, while two species, R. schlegelii and R. arboreus are distributed on the mainland, and are northernmost species of the genus and family (Matsui and Maeda, 2018). This leads us to expect the presence of genetic variations in these species induced by climatic oscillations like Cynops and Buergeria shown above.
Rhacophorus schlegelii is distributed in Honshu,Shikoku, and Kyushu Islands, and is characterized by breeding under the ground. Unlike R. arboreus, with which the species is sympatric in Honshu, it has small body size and yellow iris, and usually lacks dorsal marking. Rhacophorus arboreus occurs only in Honshu,and breeds on trees or on the ground but never in the soil. Compared with R. schlegelii, it has a large body size, sometimes with dorsal marking, and has reddish iris(Maeda and Matsui, 1999).
A few previous studies reported geographic variation of these two species. Okada and Kawano (1924) recognized two varieties of Polypedates schlegelii Günther, 1858:P. schlegelii var. arborea with a dorsal marking and P.schlegelii var. intermedia without such a marking from northern Honshu. Later, Okada (1930) elevated the two varieties to subspecies of R. schlegelii. Now R. schlegelii intermedia is synonymized with R. arboreus, which is treated as a species distinct from R. schlegelii (Nakamura and Uéno, 1963). More recently, Iizuka et al. (1990)reported geographic variation of nucleolar organizing regions in both R. schlegelii and R. arboreus. Using RFLP method, Wilkinson et al. (1996) analyzed 12S rRNA-16S rRNA of R. arboreus and reported genetic differentiation between eastern and western populations. However, no comprehensive studies have been made in either species encompassing whole distributional areas or estimating formation histories of populations.
It is expected that elucidation of historical formations of geographic variations in these two species would help understanding evolutions of their morphology and breeding habits. Additionally, it is biogeographically interesting that R. schlegelii occurs on Honshu, Shikoku,and Kyushu like many other Japanese frogs, but R.arboreus is restricted only to Honshu. We thus conducted molecular phylogenetic analyses of the two Rhacophorus species from Japan mainland to make their evolutionary histories clear.
2. Materials and Methods
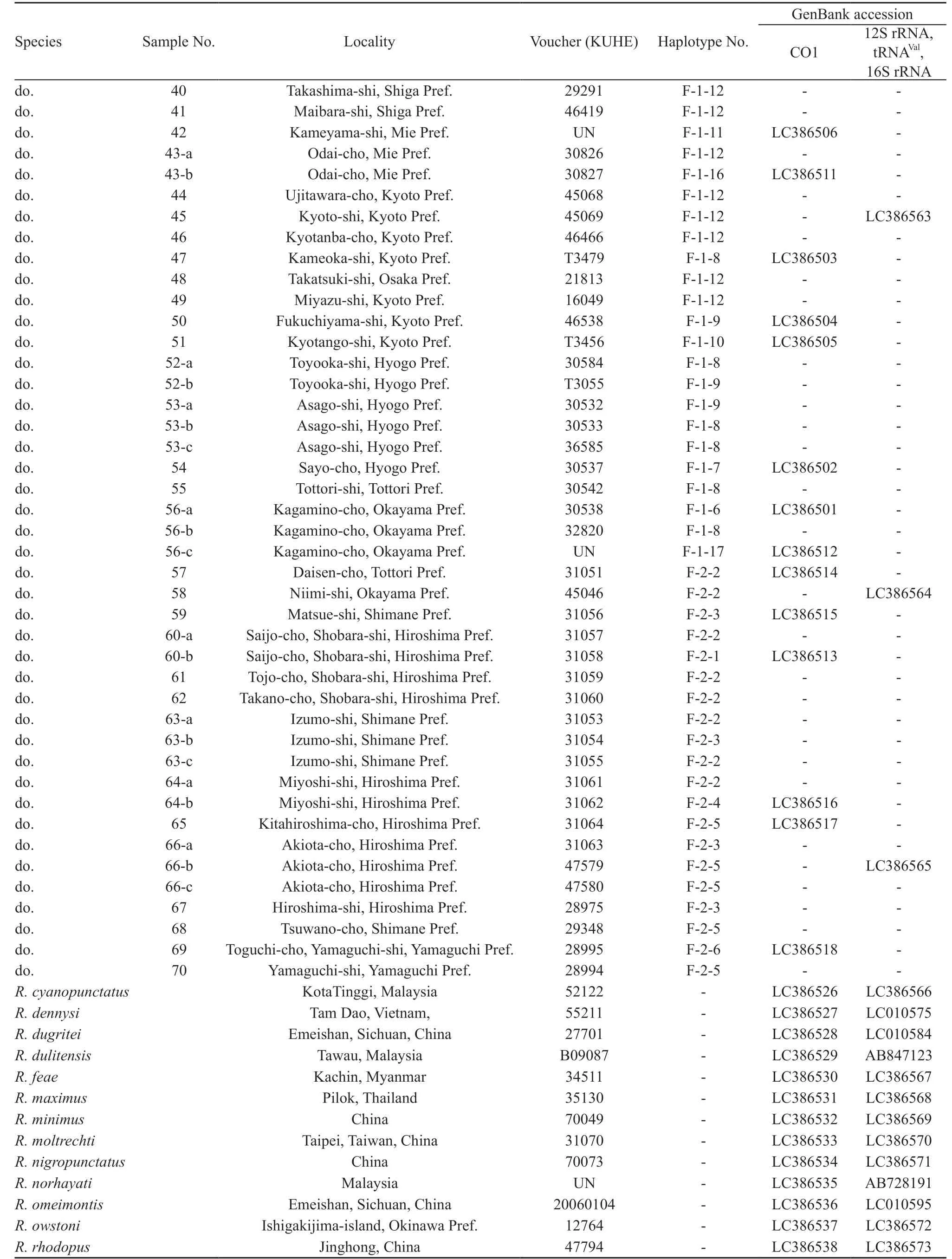
For the phylogenetic analysis between species to know the position of Japanese members among Rhacophorus,we employed a total of 17 species, including five species from Japan and 12 species from Myanmar, Thailand,Malaysia, Vietnam, and China. We determined sequences of 12S rRNA, tRNAVal, 16S rRNA, and Cytochrome C oxidase subunit 1 (CO1) regions of mitochondrial DNA for each one sample of each taxon. For outgroup, we used each one sample of Polypedates megacephalus in the subfamily Rhacophorinae, Buergeria buergeri in the subfamily Buergerinae, and Mantella madagascariensis of Mantellidae.
For the intraspecific phylogenetic analyses we determined sequences of CO1 for 229 samples of R. schlegelii from 156 localities and 95 samples of R. arboreus from 70 localities, encompassing whole ranges of their distributions (Table 1, Figure 1). For the outgroup in the analyses of R. schlegelii, R. viridis,R. amamiensis, and R. arboreus were chosen, and for those of R. arboreus, R. viridis, R. amamiensis, and R.schlegelii were chosen. Additionally, sequences of 12S rRNA, tRNAVal, and 16S rRNA were determined for R.schlegelii, using representative haplotypes obtained in the intraspecific analyses of CO1, and utilized for estimation of divergence times.
We obtained total genomic DNA from the liver or muscle samples frozen or preserved in 99% ethanol using standard phenol-chloroform extraction procedures(Hillis et al., 1996). We conducted amplifications of regions shown above by the polymerase chain reaction(PCR). The reaction conditions were initial heating at 94oC for 4 min; 33 cycles of 94oC (30 s), 53-55oC(30 s), and 72oC (2.5 min); and a final extension at 72oC for 7 min. The amplified DNA fragments were purified using polyethylene glycol (PEG, 13%). Cyclesequencing reactions were done using an ABI PRISM Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) using primers shown in Table 2. We purified the sequencing reaction products by ethanol precipitation following the manufacturer's protocol and then ran them on an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems).
We aligned sequences using the ClastalW (Thompson et al., 1994) or MAFFT version 7 (Katoh and Standley,2013), and checked the initial alignments by eye and adjusted slightly. We constructed phylogenetic trees by maximum likelihood (ML) and Bayesian (BI) methods.
The optimum substitution models for each partition were selected by Kakusan4 (Tanabe, 2011), based on the Akaike information criterion. We conducted ML analysis using TREEFINDER ver. Mar. 2011 (Jobb, 2011), and Bayesian analysis using MRBAYES v3.1.2 (Ronquist and Huelsenbeck, 2003) with the models derived from Kakusan4 based on the AIC using four simultaneous Metropolis-coupled Monte Carlo Markov chains for 7 000 000 generations and sampling a tree every 100 generations. Convergence of the log-likelihood scores was checked using TRACER ver. 1.5 (Rambaut, 2009),and discarded the first 700 000 generations as burn-in.We tested the robustness of the ML tree using bootstrap analyses (Felsenstein, 1985) with 1000 replicates. We regarded tree topologies with bootstrap values (BS) ≥70% as sufficiently resolved (Huelsenbeck and Hillis,1993) and posterior probabilities (BPP) for each branch in the Bayesian tree ≥ 95% as significant support(Huelasenbeck et al., 2001; Leaché and Reeder, 2002).We calculated genetic distances of CO1 based on mean uncorrected p-distances for pairwise combinations of clades and subclades using MEGA, version 5 (Tamura et al., 2011).
For a total 46 samples including 43 samples of the two species whose sequences of 12S rRNA, tRNAVal, 16S rRNA, and CO1 were determined, and three outgroup samples, we estimated the divergence times using a Bayesian relaxed molecular clock (Drummond et al.,2006) calculated in BEAST ver. 1.6.2 (Drummond and Rambaut, 2007) using 10 million generations, discarding the first 1 million generations as burn-in. Suitable burnin and convergence of parameters were determined using TRACER ver. 1.5 (Rambaut, 2009). As calibration points,the divergence of Buergerinae and Rhacophorinae as 45.9(95% credibility interval [CI] 57.6-36.6) million years before present (MYBP: Kim et al., 2007), of Rhacophorus and Polypedates as 30.6 (CI 34.7-25.2) MYBP (Li et al., 2013), and of Cynops ensicauda from Okinawa and Amami Islands as 5.21 (CI 6.82-3.64) MYBP (Tominaga et al., 2013) were used.
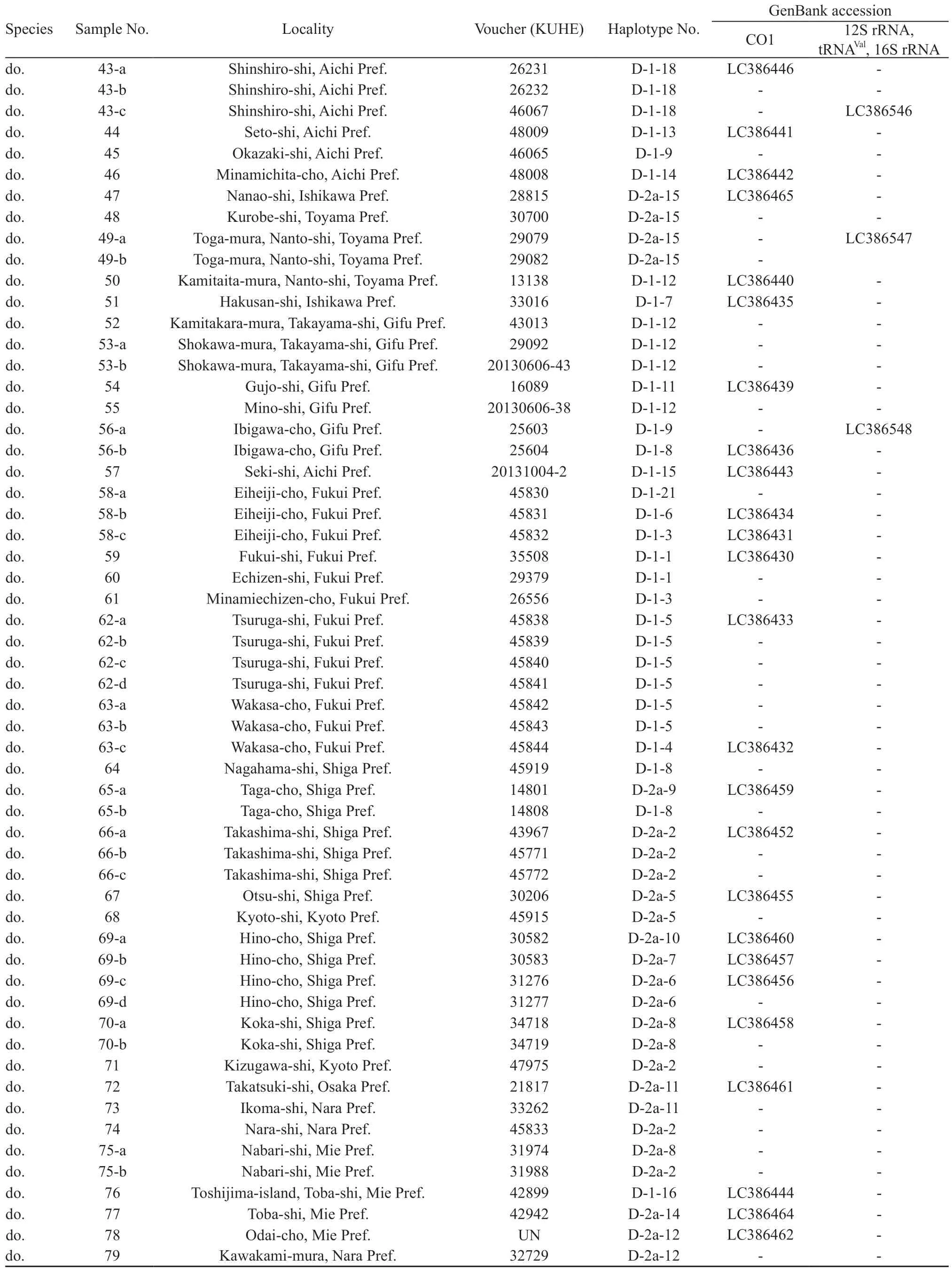
Continued Table 1
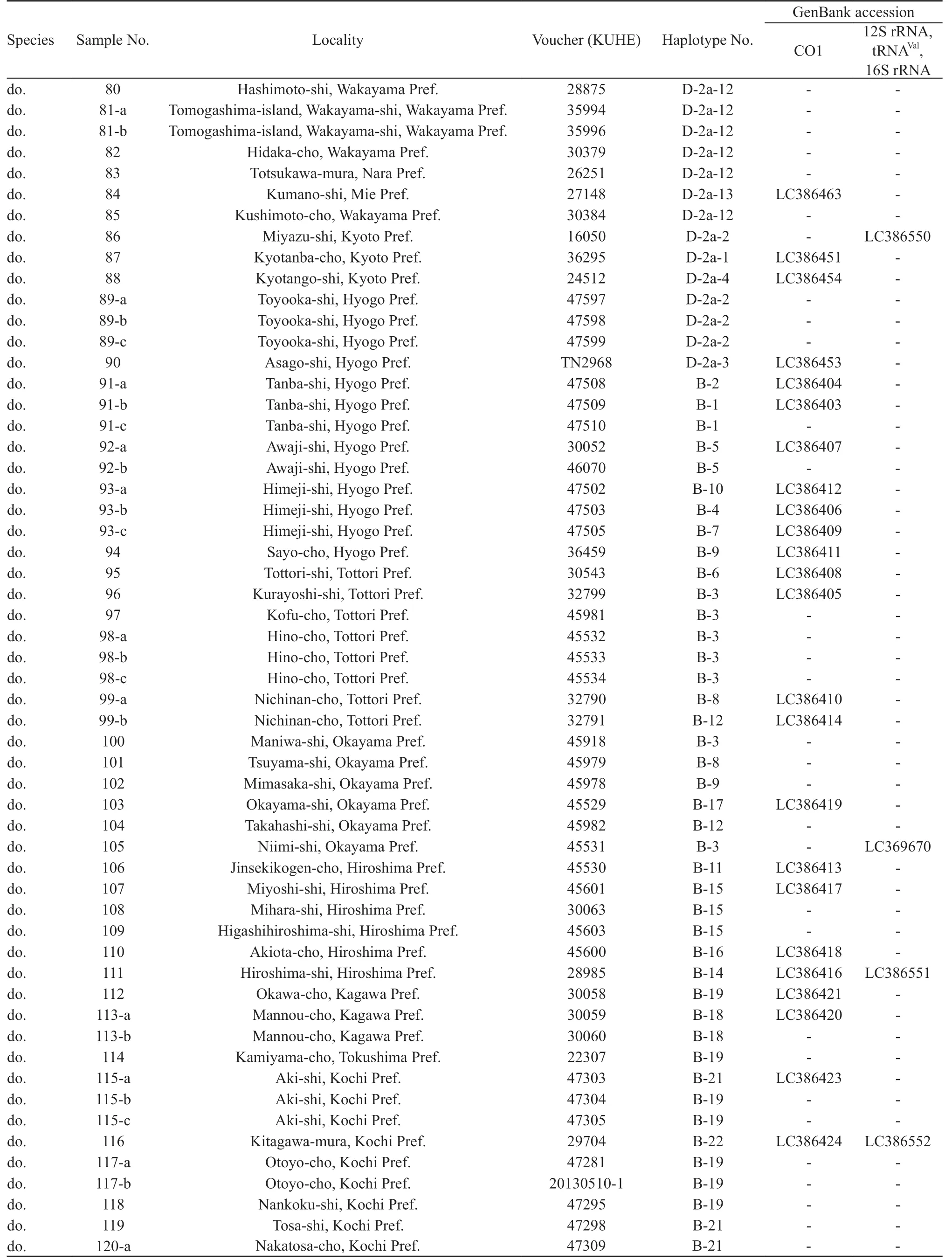
Continued Table 1
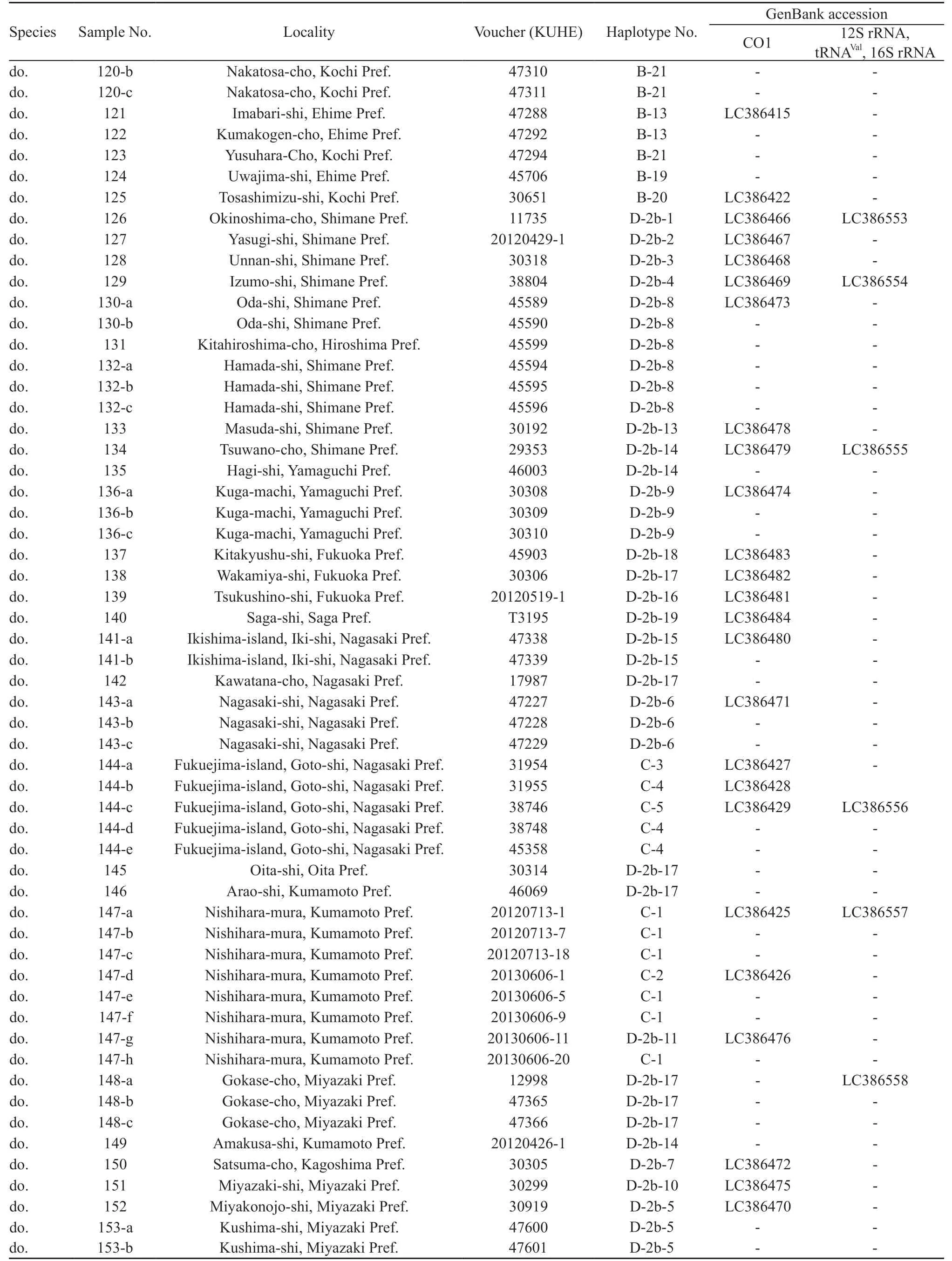
Continued Table 1
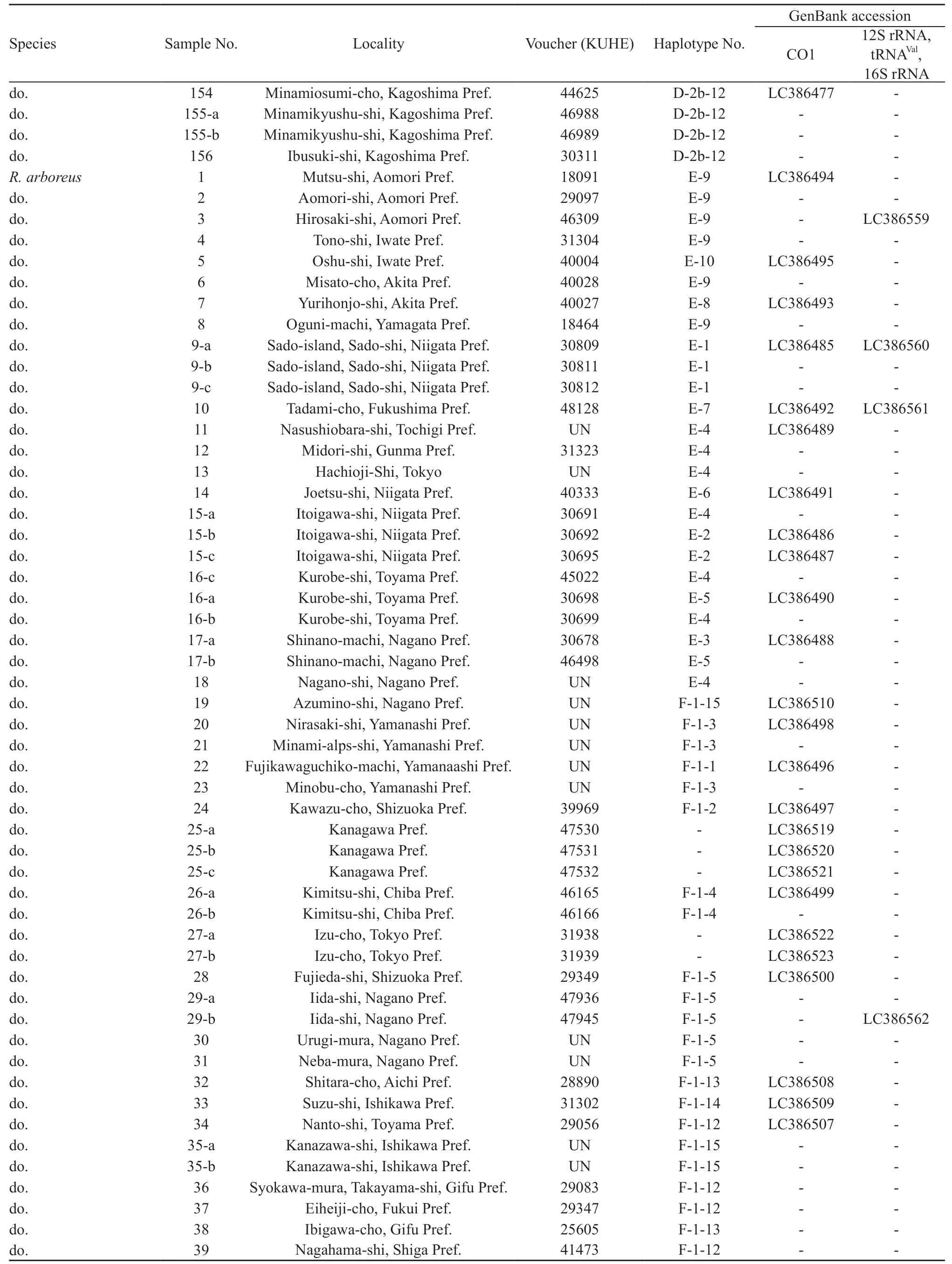
Continued Table 1
Continued Table 1
Continued Table 1
We constructed haplotype networks for a set of primers(588 bp, Rh_CO1_F1 and Rh_CO1_R1) in CO1 for R. schlegelii and R. arboreus, and for each clade and subclade, we calculated both haplotype diversity (h) and nucleotide diversity (π, based on pairwise differences)values and conducted neutrality tests. We used TCS ver. 1.21 (Clement et al., 2000) to construct haplotype networks. Statistical confidence limit was set at >95%.For R. schlegelii, network was constructed for each clade and subclade since networks were not constructed in the confidence interval of 95%. For calculation of haplotype and nucleotide diversities, and conducting neutrality tests, ARLEQUIN ver. 3.5.1.3 (Excoffier and Lischer,2010) was used. Putative populations of introduced origin(Localities 25: Kanagawa Pref. and 27: Izu-Oshima, see below in Discussion) were not included in these analyses.
3. Results
3.1. Interspecific phylogeny of RhacophorusOf 3976 nucleotide sites of 12S rRNA, tRNAVal, 16S rRNA,and CO1 regions obtained for Rhacophorus including Japanese members, 1422 were variable within the ingroup. For the ML analysis, J2 (Jobb, 2008)+G was selected by Kakusan4 as the optimal model for 12S rRNA and tRNAVal, J2 (Jobb, 2008)+G for 16S rRNA, and TN93(Tamura and Nei, 1993)+G, HKY85 (Hasegawa et al.,1985)+G, and J3 (Jobb, 2008)+G were selected as the optimal models for 1st, 2nd, and 3rd codon positions of CO1 gene. For the BI analysis, GTR+G was selected for 12S rRNA, tRNAVal, 16S rRNA, and SYM (Zharkikh,1994)+G, HKY85+G, and GTR+G were selected for 1st, 2nd, and 3rd codon positions of CO1 gene. The ML and Bayesian analyses produced topologies with -ln L =21 917.505 and 21 918.380, respectively.
Phylogenetic analyses employing two different optimality criteria yielded nearly identical topologies.As shown in the ML tree (Figure 2), Japanese and some Chinese species formed a clade (ML BS = 99%, BPP= 1.00) in which R. owstoni and R. moltrechti formed a clade (100%, 1.00), which was sister to the clade(70%, 1.00) of other Japanese taxa and Chinese species.However, other than the monophyly of R. viridis and R.amamiensis, phylogenetic relationships within Japanese Rhacophorus were not resolved.
3.2. Intraspecific phylogeny of R. schlegeliiFrom 1554 nucleotide sites of complete CO1 region obtained for 229 individuals of R. schlegelii, 150 haplotypes were recognized, and 420 positions were found variable including the outgroup. For the ML analysis, Kakusan4 selected TN93+G, HKY85, and TN93+G as the optimal models for 1st, 2nd, and 3rd codon positions of CO1 gene, respectively. For the BI analysis, SYM+G, F81(Felsenstein, 1981)+G, and GTR+G, were selected respectively, for each position
Phylogenetic analyses yielded identical topologies in the two different criteria, and only the ML tree is shown(Figure 3A). Rhacophorus schlegelii formed a clade with high supports (99%/1.00) in which four major clades,each with high support, were recognized (A: 99%/1.00;B: 99%/1.00; C: 98%/1.00; D: 93%/1.00), although their relationships were unresolved. In addition, three subclades were recognized within Clade D (D-1: 99%/1.00; D-2a:99%/1.00; D-2b: 100%/1.00).
In the phylogenetic trees obtained for 12S rRNA,tRNAVal, 16S rRNA, and CO1, all the four clades obtained in the CO1 trees were recognized, and Clade A proved to be a sister clade to the clade of the remaining three clades(87%/0.99), each of which formed a clade (B: 99%/1.00;C: 99%/1.00; D: 98%/1.00), but their relationships were unresolved.
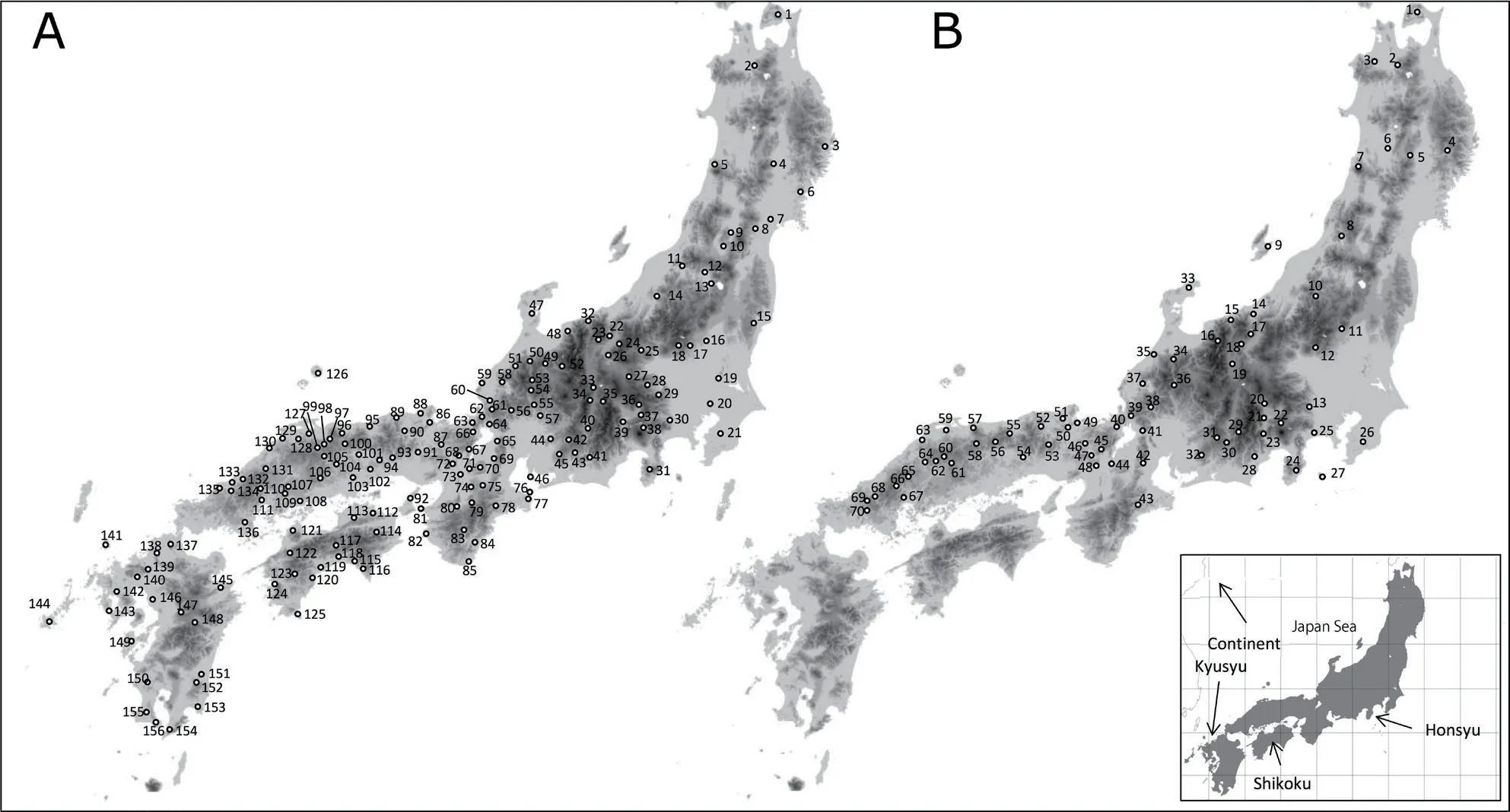
Figure 1 Map of sampling localities of (A) Rhacophorus schlegelii and (B) R. arboreus. For locality name, see Table 1.
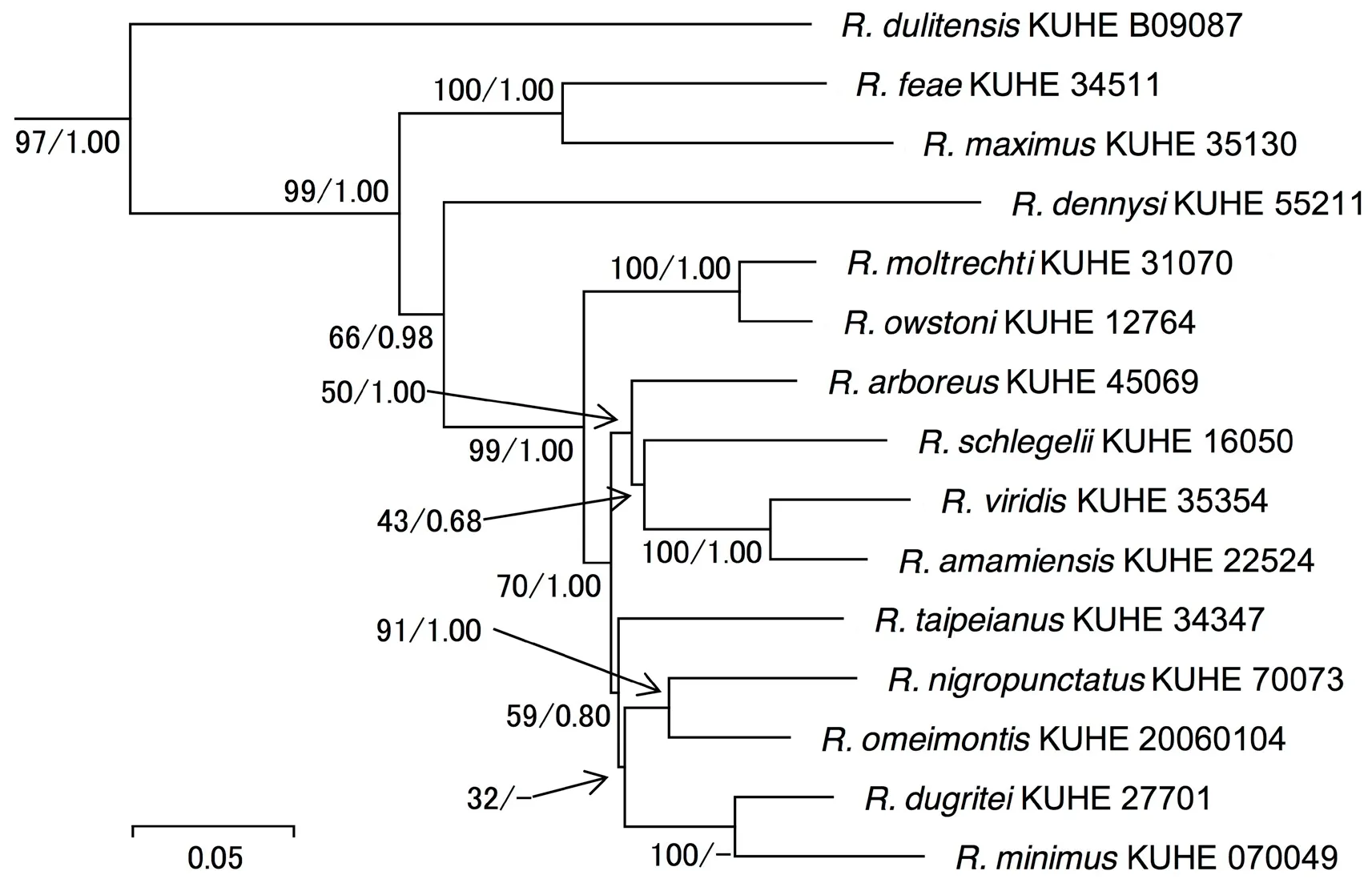
Figure 2 Maximum likelihood (ML) tree based on mitochondrial 12S rRNA, tRNAVal, 16S rRNA and CO1 genes of the genus Rhacophorus.Nodal values indicate bootstrap for ML and Bayesian posterior supports (ML-BS/BSS). For voucher number and DDBJ accession number,see Table 1.
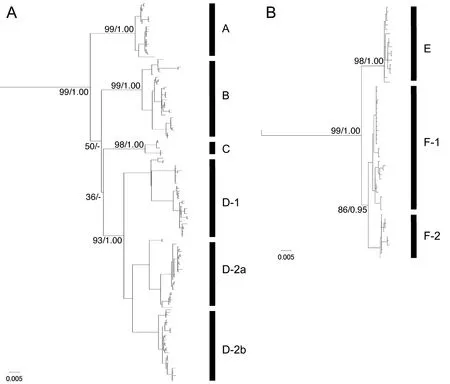
Figure 3 Maximum likelihood (ML) tree based on mitochondrial CO1 gene of (A) R. schlegelii and (B) R. arboreus. Nodal values indicate bootstrap supports for ML and Bayesian posterior probably (ML-BS/BSS). For locality numbers, see Figure 1 and Table 1.
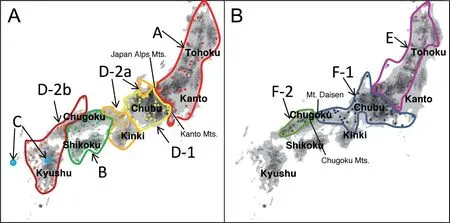
Figure 4 Geographic distribution of each clade and subclade of (A) R. schlegelii and (B) R. arboreus. For sample marks and lines, refer to Figure 1.
As shown in Figure 4A, range of distribution was Tohoku, Kanto, and eastern Chubu regions of Honshu Island in Clade A, westernmost Kinki and eastern Chugoku regions of Honshu, and Shikoku Island in Clade B, and a part of Kyushu Island (localities 144 and 147) in Clade C. Clade D had wider range than others in Chubu, Kinki, western Chugoku and Kyushu regions,with Subclade D-1 in Chubu region except for north and Toshijima Island (locality 76), Subclade D-2a in northern Chubu and Kinki regions, and Subclade D-2b in western Chugoku and Kyushu regions. Of these, sympatric distribution was found between Clades C and D (at locality 147) and Subclades D-1 and D-2a (at locality 65).Other clades and subclades occur parapatrically (Figure 4A).
Mean (± 1SE, in %) of uncorrected p-distance was 5.09 ± 0.51 between Clades A and B, 4.61 ± 0.50 between Clades A and C, 5.36 ± 0.48 between Clades A and D,4.70 ± 0.47 between Clades B and C, 4.93 ± 0.39 between Clades B and D, and 4.80 ± 0.43 between Clades C and D. In Clade D, they were 4.56 ± 0.48 between Subclades D-1 and D-2a, 3.69 ± 0.39 between D-1 and D-2b, and 3.30 ± 0.41 between D-2a and D-2b.
3.3. Intraspecific phylogeny of R. arboreusIn R.arboreus, 58 haplotypes were recognized within 95 individuals. Of 1 554 bp of complete CO1 region, 347 positions were found variable. For the ML analysis,Kakusan4 selected TN93+G, HKY85, and J1 (Jobb,2008)+G, respectively, as the optimal models for 1st,2nd, and 3rd codon positions of CO1 gene. For the BI analysis, SYM+G, F81+G, and GTR+G, were selected respectively, for each position.
The ML and Bayesian analyses produced topologies with -ln L = 4 240.711 and 4 247.162, respectively.Identical topologies were obtained in ML and BI, and ML tree is shown in Figure 3B. Rhacophorus arboreus formed a clade with high supports (99%/1.00), within which two major clades, Clade E (98%/1.00) and Clade F (86/0.95),were recognized, and two Subclades (F-1: 78%/1.00 and F-2: 98%/1.00) were recognized in the latter.
Clade E occurs in Tohoku, Kanto except for southern part, and eastern Chubu regions, and Clade F in southern Kanto, Chubu, Kinki, and Chugoku regions (Figure 4B). In Clade F, Subclade F-1 is found in southern Kanto, Chubu, and Kinki regions, and Subclade F-2 in Chugoku regions. These clades and subclades occurred allopatrically. Topotypic samples of synonymized subspecies R. schlegelii intermedia (locality 9) were included in Subclade F-1 without remarkable genetic differentiation among R. arboreus from the surrounding area.
Mean (± 1SE, in %) of uncorrected p-distance was 2.40± 0.33 between Clades E and F, and 1.46 ± 0.24 between F-1 and F-2.
3.4. Divergence timeRhacophorus owstoni formed a clade with R. moltrechti in Taiwan, China and split from the group containing other Japanese Rhacophorus at 7.68 (9.44-6.05) MYBP. Rhacophorus owstoni and R.moltrechti were estimated to have diverged at 2.32 (3.35-1.47) MYBP. Japanese members other than R. owstoni were estimated to have split from the common ancestor of Chinese species including R. omeimontis and R. minimus at 7.01 (8.53-5.51) MYBP. Divergence of R. viridis and R. amamiensis was estimated to be 3.57 (4.56-2.58)MYBP.
Within R. schlegelii, Clade A and others first diverged at 2.40 (3.02-1.79) MYBP, and divergences among Clades B, C, and D occurred after 2.04 (2.57-1.52)MYBP. Differentiation within each clade and subclade was estimated to be 0.51 (0.71-0.32) MYBP in Clade A, 0.84 (1.15-0.55) MYBP in CladeB, 0.52 (0.79-0.27)MYBP in Clade C, 1.44 (1.80-1.05) MYBP between Subclade D-1 and others in Clade D, and 1.20 (1.53-0.87)MYBP between Subclades D-2a and D-2b. Differentiation within Subclade D-1 was 0.76 (1.02-0.50) MYBP, D-2a was 0.66 (0.90-0.42) MYBP, and D-2b was 0.44 (0.64-0.25) MYBP.
Within R. arboreus, divergence of Clades E and F was estimated to have occurred at 0.96 (1.30-0.65) MYBP.Within each clade or subclade, age of differentiation was estimated to be 0.23 (0.35-0.12) MYBP in Clade E, 0.65(0.90-0.42) MYBP in Clade F, 0.30 (0.46-0.14) MYBP in Subclade F-1, and 0.19 (0.30-0.08) MYBP in Subclade F-2.
3.5. Haplotype network and population genetic analysisHaplotype network of R. schlegelii, based on 588 bp of CO1 region of each clade and subclade is shown in Figure 5A, and haplotype diversity, nucleotide diversity, and results of neutrality tests (Fu's FS: Fu,1997) are shown in Table 3. Clade A tended to have lower nucleotide diversity than others, and Clades A and C tended to have lower haplotype diversity than others. Fu's FS were significantly negative (P < 0.05) in Clades A and D, and Subclade D-2b.
Haplotype network of R. arboreus, based on 588 bp of CO1 region of each clade and subclade is shown in Figure 5B, and haplotype diversity, nucleotide diversity,and result of test of neutrality (Fu's FS) are shown in Table 3. Mean of haplotype diversities (0.8646) was slightly smaller and nucleotide diversity (0.0043) was much smaller than in R. schlegelii (0.8981 and 0.0124,respectively). In the two clades recognized (E and F) and Subclade F-1, Fu's FS were significantly negative, but in Subclade F-2, the values were not significantly negative.
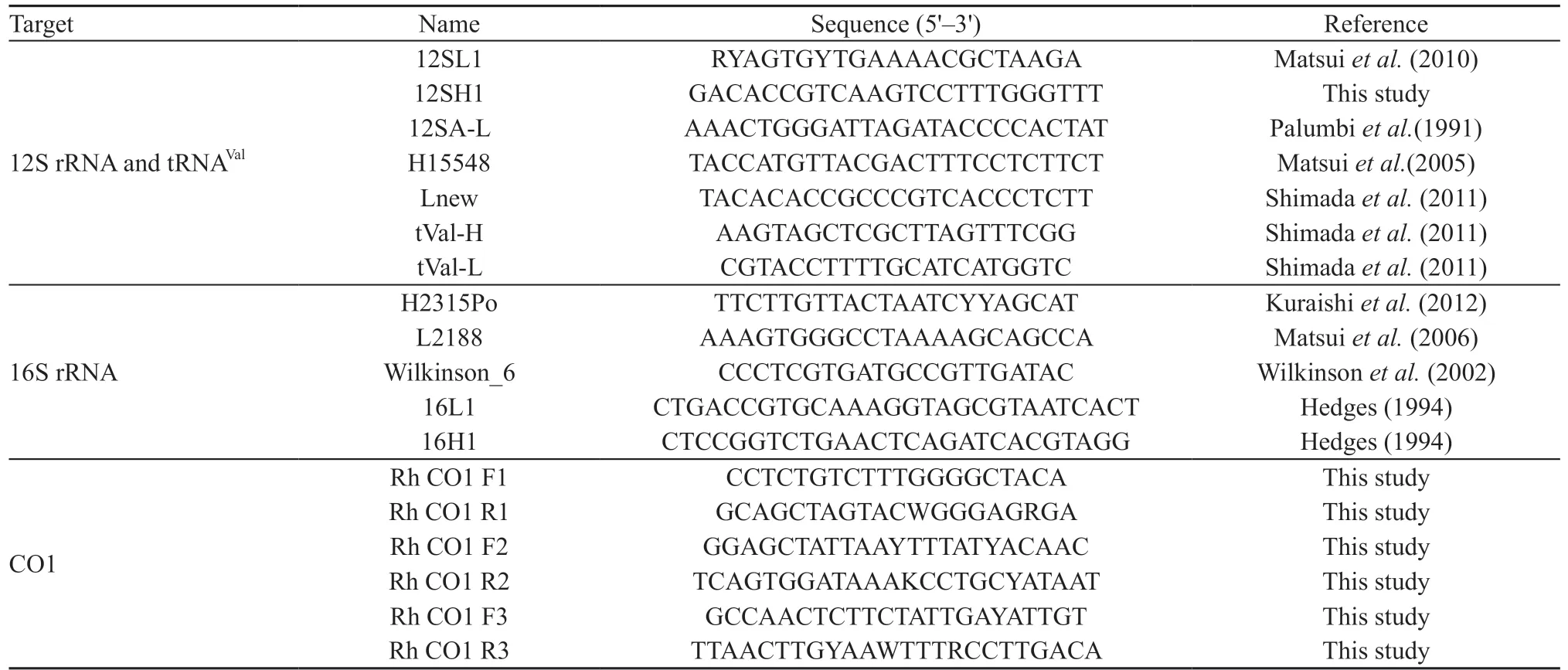
Table 2 Primers used to amplify mtDNA in this study.
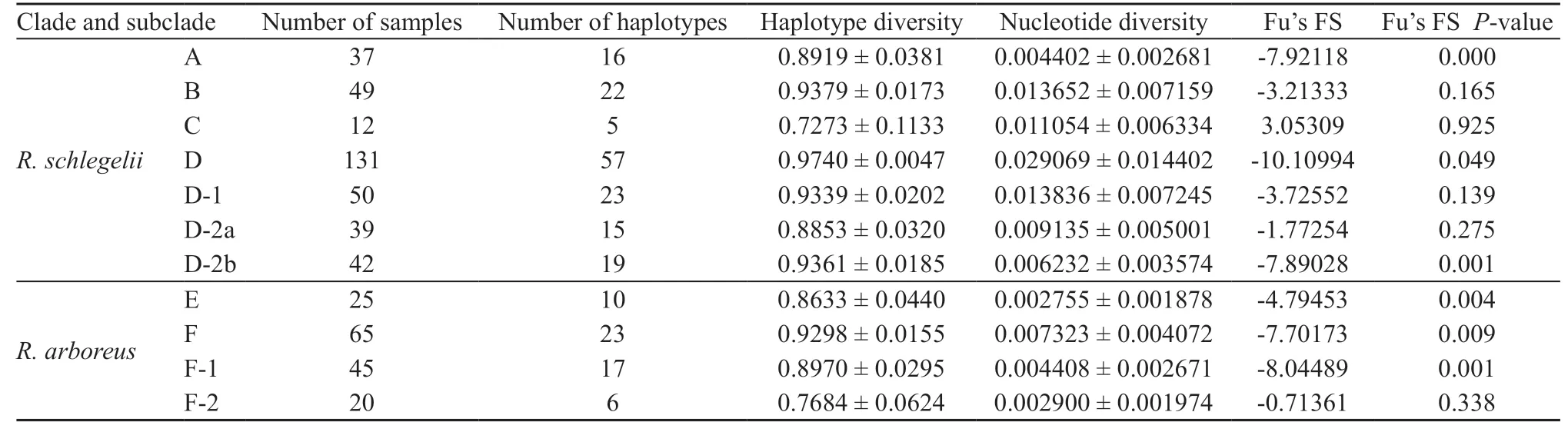
Table 3 Genetic diversity indices and result of Fu's tests of neutrality (Fu, 1997).
4. Discussion
4.1. Phylogenetic relationships of Japanese RhacophorusLi et al. (2013) and Hertwig et al. (2013)similarly recognized three clades (I: mainly Bornean species; II: flying frogs; III: species occurring mainly in East Asia) in the genus Rhacophorus. Our result corroborated them and all Japanese Rhacophorus are included in the group mainly from East Asia (Clade III),which formed a monophyletic group against Clade I (R.cyanopunctatus and R. gauni) and Clade II (R. norhayati and R. rhodopus). Within Clade III of mainly East Asian species group, southern species like R. dulitensis, R.feae, and R. maximus diverged first and Japanese species positioned relatively terminally on the phylogram. From this relationship, species in this group are estimated to have originated in the south and sometime later invaded north to form present distribution.
It is difficult to elucidate interspecific phylogenetic relationships among Rhacophorus, but among Japanese taxa, R. owstoni from the Yaeyama group of the southern Ryukyus formed a clade with R. moltrechti from Taiwan,China and split from the remaining Japanese and Chinese species. Except for R. owstoni, Japanese Rhacophorus had ancestor common to species like R. taipeianus in Taiwan,China and Chinese R. omeimontis, R. dugritei, and R.minimus, although their relationships were unresolved. It was estimated that the differentiation of these frogs began 7.01 MYBP, which is much older than the period of last disjunction of Ryukyu Archipelago and mainland Japan(1.70 MYBP: Kitamura and Kimoto, 2004). From this it is estimated that divergence of ancestors of Japan and the continent already occurred by unknown factors before the isolation of Japanese Islands and Ryukyu Archipelago from continental China by oceans.
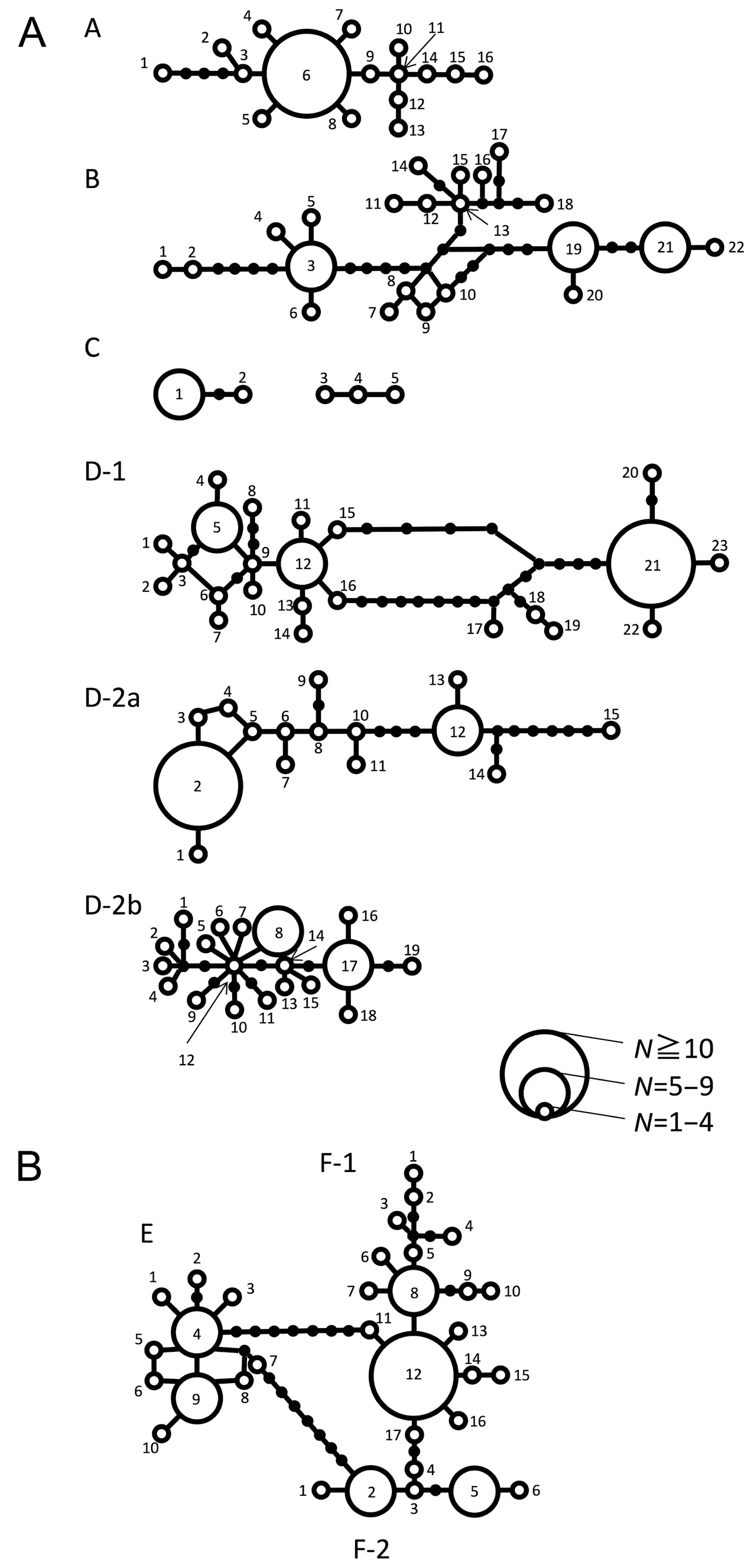
Figure 5 Statistical parsimony network of each clade and subclade of (A) R. schlegelii and (B) R. arboreus. Filled circles indicate missing haplotypes. The size of each open circle is proportional to the haplotype frequency.
4.2. Phylogenetic relationships in R. schlegeliiOn the phylogenetic tree based on multiple genes, R. schlegelii was split into four clades. Among them Clade A was the first to diverge from the others, and therefore, R.schlegelii was divided into two groups roughly from eastern and western regions (Figure 4A).
Near the boundaries of these clades, Japan Alps and Kanto Mountains are situated between Clades A and D, and Chugoku Mountains between Clades B and D.Current boundary of Clades A and D is positioned in the east of Japan Alps, but the boundary is estimated to have been more west in the past since the haplotype of Clade A occurs isolated on the Izu Peninsula. Clade A and the other clades were estimated to have diverged around 2.40 MYBP. It is estimated that the altitude of Hida Mountains(northern Japan Alps) was ca. 2000 m but of Akaishi Mountains (southern Japan Alps) was ca. 1000 m in that era (Chinzei and Machida, 2001). Therefore, it is not likely that Akaishi Mountains at that era were a barrier for the migration of R. schlegelii, which mainly inhabits from lowland to subalpine regions. However, around 2.70 MYBP, which nearly corresponds to the age of divergence between Clade A and the others (2.40 MYBP), cycles of the glacial and the interglacial periods are estimated to become evident (Mix et al., 1995), and climatic factors like lowering of air temperatures on high mountains would have inhibited population gene exchange and promoted the differentiation of R. schlegelii.
Boundary of Clades B and D is roughly situated near Chugoku Mountains. On the Japan Sea side, Clade B is distributed between Subclades D-2a and D-2b. In the past,Subclades D-2a and D-2b are estimated to have occurred in the north, while Clade B occurred in the south, of Chugoku Mountains. Subsequently, Clade B is estimated to have widened the range locally in the north of Chugoku Mountains. It is estimated that Subclades D-2a and D-2b first differentiated and Clade B later was distributed at their boundary, since differentiation within Clade B was 0.84 MYBP, while divergence of Subclades D-2a and D-2b was 1.20 MYBP. It is possible that divergence of Subclades D-2a and D-2b was affected by the presence of active volcano of Mt. Daisen and cooler climate of Japan Sea side, but the volcanic activity of Daisen is estimated to have begun 0.90 MYBP (Ota et al., 2004),and there seems to have been no significant climatic variation. Furthermore, altitudes of Chugoku Mountains are at most 800-1000 m a.s.l. (Ota et al., 2010), which is hardly considered to be a barrier of distribution, inducing differentiations of R. schlegelii. Thus the reasons for differentiations of Clades B and D, and of Subclades D-2a and D-2b are unknown.
Clades C and D occurred sympatrically at the location 147 (Kumamoto, western central Kyushu). It is conceivable that Clade D invaded to the range of Clade C in Kyushu and occupied present areas.
Differentiations within Clade A were less marked than in other clades, and haplotype diversity and nucleotide diversity were smaller than in Clades B and D (Table 3). Additionally, results of neutrality tests indicated past rapid expansion of distribution range, and the haplotype exhibited star-like network (Figure 5A), with the commonest haplotype (A-6) occurring in Kanto and eastern Chubu districts, indicating that Clade A utilized these regions as refugia in the past (Figure 6A).
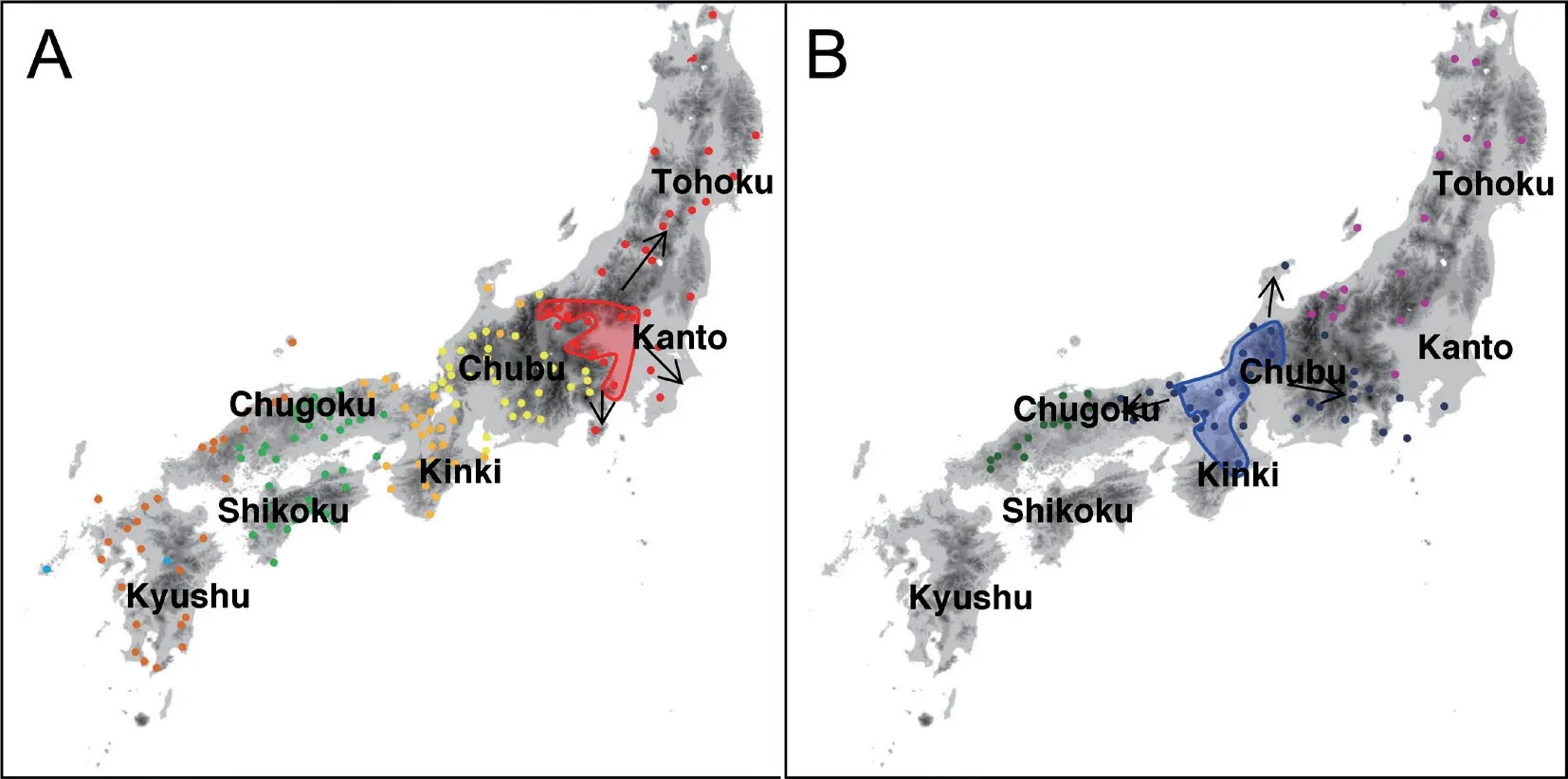
Figure 6 Examples of dispersals in (A) R. schlegelii (Clade A [A-6]) and (B) R. arboreus (Subclade F-1 [F-1-12]) that were haplotypes most frequently observed.
Degree of genetic differentiation in Clade B, as shown by haplotype diversity and nucleotide diversity was smaller than Clade D, which includes diversified subclades, but was larger than in Clades A and C (Table 3). Haplotype network contains long branches among each haplotype (Figure 3). This indicates that Clade B has originated older than Clade A but younger than Clade D,and sustained population size without experiencing largescale changes in distribution.
Clade C from two distant localities in Kyushu had lower haplotype diversity than in other clades, but had nucleotide diversity higher than Clade A, Subclades D-2a and D-2b (Table 3). Divergence time between disjunct populations was 0.52 MYBP, and are estimated have been distributed widely on Kyushu at least by that period.Probably, the clade was substituted by Subclade D-2b,which invaded after Clade C decreased the population size through volcanic activities in northern Kyushu (2.0-0.5 MYBP: Sakamoto, 2009). Occurrence of genetically different lineages in Kyushu is known in the newt Cynops pyrrhogaster (Tominaga et al., 2013) and the brown frog Rana tagoi (Eto et al., 2012), and Clade C might have experienced geohistrical events similar to them.
Clade D showed larger genetic differentiation than in other clades, and three fairly divergent subclades were recognized. Of these, Subclade D-2b was estimated to have experienced rapid range expansion based on neutrality test (Table 3). The subclade began differentiation at 0.44 MYBP, and is estimated to have occurred in westernmost Honshu and northern Kyushu,because the dominant haplotypes (D-2b-8, 17) are obtained from these areas (Figure 5, Table 1). Then,expansion of distribution towards southern Kyushu on Subclade D-2b seems to have induced restricted distribution of Clade C shown above.
4.3. Phylogenetic relationships in R. arboreus
Phylogenetic analyses using CO1 gene revealed the presence of eastern Clade E and western Clade F in R. arboreus, with further two subclades in Clade F(Figures 3B and 4B). These clades and subclades occur parapatrically, and the boundary of Clades E and F was near the region connecting Japan Alps and Kanto Mountains. However, because no differentiation is found between many populations with mountains or mountain ranges in between, Japan Alps and Kanto Mountains are hardly considered to have induced genetic differentiation.The degree of genetic differentiation is much smaller than in R. schlegelii: mean uncorrected p-distance was 2.4%between Clades E and F of R. arboreus, as compared with 5.4% between Clades A and D of R. schlegelii. Thus, R.arboreus might have experienced events where they lost genetic diversities, and retreated to refugia, indicating genetic differentiations in R. arboreus to have been induced by their retreats to refugia.
Clade E from eastern Japan had lower degree of genetic differentiation, haplotype diversity, and nucleotide diversity than western Clade F (Table 3), and was suggested to have experienced past rapid expansion of distribution range based on the neutrality test. In the range of Clade E, haplotypes E-4 in Tohoku and E-9 in northern Chubu were found in high frequency and were distributed widely. This suggests that the original populations of these haplotypes that started their rapid range expansions were present at their boundaries, which would have been their refugia.
Within Clade F, the Subclade F-1 showed significant negative value in the neutrality tests and suggested past rapid range expansion (Table 3). Within Subclade F-1,the haplotype F-1-12 was found frequently, and exhibited star-like haplotype network (Figure 5B). This haplotype occurred widely from Kinki to northern Chubu regions,and might utilize this area as their refugia (Figure 6B).
No evidence of range expansion or reduction was obtained in the Subclade F-2, and differentiation in the subclade was 0.19 MYBP, in contrast to 0.65 MYBP estimated for the differentiation from Subclade F-1.The haplotype network exhibits array-like pattern. This indicates that the Subclade F-2 was already distributed in the Chugoku Mountains in 0.65 MYBP and sustained stable population size and distributional range. In this way, R. arboreus seems to have experienced to retreat to the refugia, and caused genetic differentiations among local populations thus formed. It is estimated that the populations in central Honshu retreated to northern regions, as if avoiding warm climates, but no evidence was found for such a hypothesis. Retreats to refugia might relate to the limited distribution of R. arboreus to Honshu,but we could not get any evidence for that.
Localities 25 (Kanagawa Pref.) and 27 (Izu-Oshima)were isolated from other localities in Clade F. Of these,locality 27 is represented by artificially introduced populations (Maeda and Matsui, 1999), and locality 25, with the haplotype same as Kinki region, is also considered an alien population.
4.4. Comparative phylogeographyThis study clarified that both R. schlegelii (Clade A and others) and R.arboreus (Clades E and F) are genetically divergent between the east and west regions of Japan. Such eastern and western genetic divergence is known in several wide-ranging Japanese frogs, although the boundaries are variable (e.g., Bufo japonicus [Igawa et al., 2006];Hyla japonica [Dufresnes et al., 2016]; Rana japonica[Sumida and Ogata, 1998]). The east-west boundary of R. schlegelii positioned in the east of Japan Alps, which is common to R. arboreus, except for the distribution of Clade F in Chubu and southern Kanto regions.Additionally, the boundary of Subclades D-2a and D-2b in R. schlegelii positioned in Chugoku region, like the boundary of Subclades F-1 and F-2 in R. arboreus. In this way, boundaries of distributions between genetic groups corresponded well in the two Rhacophorus species. However, estimated ages of differentiations greatly differed between the two species and their current distributions are hardly attributable to identical geological events or geographic boundaries.
Both the two species of Rhacophorus were suggested to include genetic populations that experienced retreatment to refugia. Among them, typical cases are as follows: in R. schlegelii, populations now occurring in Tohoku, Kanto, and eastern Chubu regions (Clade A)are estimated to have utilized Kanto and eastern Chubu regions as their refugia, while in R. arboreus, populations occurring Chubu to Kinki regions (Subclade F-1) are estimated to have refugia in the northern part of that regions. In addition, Subclade D-2b in R. schlegelii and Clade E in R. arboreus indicate the trace of retreat to refugia.
Rhacophorus arboreus was found much less diversified genetically than R. schlegelii, although it is more variable than the latter species in morphology and breeding habits (Nakamura and Uéno, 1963; Matsui and Maeda, 2018). At present, Clade A of R. schlegelii and Clade E of R. arboreus occur sympatrically, but because R. schlegelii is estimated to have retreated to Kanto and eastern Chubu regions, wide areas of eastern Japan is estimated to be occupied only by R. arboreus. In western Japan, R. arboreus and R. schlegelii are estimated to have coexisted long because R. arboreus retreated to north of Kinki and Chubu regions. In the regions where the two species co-occurred, they interfered each other,which might have resulted in enhancement of the habit of breeding on trees and possession of body markings in the western populations of R. arboreus. In order to test this hypothesis, knowledge of morphology and breeding habits, both now considered not variable, of R. schlegelii should be accumulated and interspecific interference at the sympatric zone should be studied ecologically.
AcknowledgmentsWe thank M. AMANO, K.B. DONTCHEV, M. HASEGAWA, T. HAYASHI,T. HIKIDA, S. HONJYO, T. KAKINOKI, K.KASUGAI, J.-B. KIM, K. KOMIZO, T. KUBOTA, J.MARUNOUCHI, MM. MATSUI, T. MATSUKI, K.MATSUO, Y. MISAWA, Y. MIYAGATA, K. MOCHIDA,A. MORI, S. MORI, T. NAKANO, T. NISHIMURA, S.OKADA, T. OKAMOTO, A. SAKABE, A. SAKAI, M.SAKAMOTO, T. SASAKI, T. SHIMADA, Z. SHIMIZU,R. SHIMOYAMA, T. SUGAHARA, T. SUGIHARA, S.TANABE, A. TOMINAGA, Y. UEDA, A. YAMADA,M. YAMAGAMI, A. YAMANE, Y. YASUKAWA, Y.YUASA, and M. YUMOTO for help in collecting sample material, and G. AOKI, A. HAMIDY, N. KURAISHI,and N. YOSHIKAWA for laboratory assistance. The research was partly supported by Grants-in-Aid from the Monbukagakusho through the JSPS (Nos. 10041166,20405013, and 23405014) and by a grant from The U. S.National Geographic Society (No. 4505-91) to MM.
 Asian Herpetological Research2019年2期
Asian Herpetological Research2019年2期
- Asian Herpetological Research的其它文章
- Genetic Structure of the Red-spotted Tokay Gecko, Gekko gecko(Linnaeus, 1758) (Squamata: Gekkonidae) from Mainland Southeast Asia
- Comparative Skin Histology of Frogs Reveals High-elevation Adaptation of the Tibetan Nanorana parkeri
- On the Generic Taxonomy of Opisthotropis balteata (Cope, 1895)(Squamata: Colubridae: Natricinae): Taxonomic Revision of Two Natricine Genera
- Evaluating the Importance of Environmental Variables on Spatial Distribution of Caspian cobra Naja oxiana (Eichwald,1831) in Iran
- Appendix 1 Specimens examined.
